6O7K
 
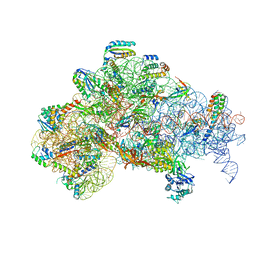 | | 30S initiation complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Frank, J, Gonzalez Jr, R.L, kaledhonkar, S, Fu, Z, Caban, K, Li, W, Chen, B, Sun, M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-29 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Late steps in bacterial translation initiation visualized using time-resolved cryo-EM.
Nature, 570, 2019
|
|
5CE7
 
 | | Structure of a non-canonical CID of Ctk3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CTD kinase subunit gamma | | Authors: | Muehlbacher, W, Mayer, A, Sun, M, Remmert, M, Cheung, A.C, Niesser, J, Soeding, J, Cramer, P. | | Deposit date: | 2015-07-06 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Ctk3, a subunit of the RNA polymerase II CTD kinase complex, reveals a noncanonical CTD-interacting domain fold.
Proteins, 83, 2015
|
|
5GHE
 
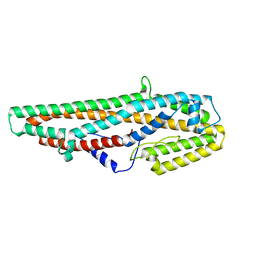 | | Crystal Structure of Bacillus thuringiensis Cry6Aa2 Protoxin | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Huang, J, Guan, Z, Zou, T, Sun, M, Yin, P. | | Deposit date: | 2016-06-19 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal structure of Cry6Aa: A novel nematicidal ClyA-type alpha-pore-forming toxin from Bacillus thuringiensis
Biochem.Biophys.Res.Commun., 478, 2016
|
|
3U1R
 
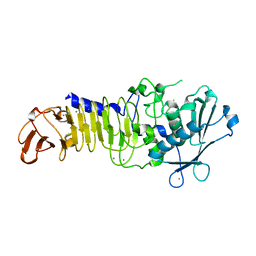 | | Structure Analysis of A New Psychrophilic Marine Protease | | Descriptor: | Alkaline metalloprotease, CALCIUM ION, ZINC ION | | Authors: | Zhang, S.-C, Sun, M, Tang, L, Wang, Q.-H, Hao, J.-H, Han, Y, Hu, X.-J, Zhou, M, Lin, S.-X. | | Deposit date: | 2011-09-30 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure analysis of a new psychrophilic marine protease.
Plos One, 6, 2011
|
|
4PKM
 
 | | Crystal Structure of Bacillus thuringiensis Cry51Aa1 Protoxin at 1.65 Angstroms Resolution | | Descriptor: | Cry51Aa1, GLYCEROL, GLYCINE, ... | | Authors: | Xu, C, Chinte, U, Chen, L, Yao, Q, Zhou, D, Meng, Y, Li, L, Rose, J, Bi, L.J, Yu, Z, Sun, M, Wang, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Cry51Aa1: A potential novel insecticidal aerolysin-type beta-pore-forming toxin from Bacillus thuringiensis.
Biochem.Biophys.Res.Commun., 462, 2015
|
|
1YT9
 
 | | HIV Protease with oximinoarylsulfonamide bound | | Descriptor: | (S)-N-((2S,3R)-3-HYDROXY-4-(4-((E)-(HYDROXYIMINO)METHYL)-N-ISOBUTYLPHENYLSULFONAMIDO)-1-PHENYLBUTAN-2-YL)-3-METHYL-2-(3 -((2-METHYLTHIAZOL-4-YL)METHYL)-2-OXOIMIDAZOLIDIN-1-YL)BUTANAMIDE, Pol polyprotein | | Authors: | Yeung, C.M, Klein, L.L, Flentge, C.A, Randolph, J.T, Zhao, C, Sun, M, Dekhtyar, T, Stoll, V.S, Kempf, D.J. | | Deposit date: | 2005-02-10 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Oximinoarylsulfonamides as potent HIV protease inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4F
 
 | | Synthesis and Activity of N-Axyl Azacyclic Urea HIV-1 Protease Inhibitors with High Potency Against Multiple Drug Resistant Viral Strains. | | Descriptor: | (5R,6R)-5-BENZYL-6-HYDROXY-2,4-BIS(4-HYDROXY-3-METHOXYBENZYL)-1-[3-(4-HYDROXYPHENYL)PROPANOYL]-1,2,4-TRIAZEPAN-3-ONE, Pol polyprotein | | Authors: | Zhao, C, Sham, H, Sun, M, Lin, S, Stoll, V, Stewart, K.D, Mo, H, Vasavanonda, S, Saldivar, A, McDonald, E. | | Deposit date: | 2005-06-28 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and activity of N-acyl azacyclic urea HIV-1 protease inhibitors with high potency against multiple drug resistant viral strains
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2EX0
 
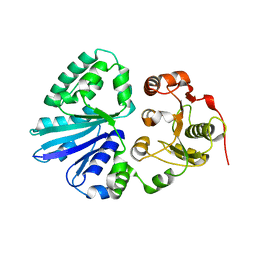 | | Crystal structure of multifunctional sialyltransferase from Pasteurella Multocida | | Descriptor: | a2,3-sialyltransferase, a2,6-sialyltransferase | | Authors: | Ni, L, Sun, M, Chen, X, Fisher, A.J. | | Deposit date: | 2005-11-07 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cytidine 5'-Monophosphate (CMP)-Induced Structural Changes in a Multifunctional Sialyltransferase from Pasteurella multocida
Biochemistry, 45, 2006
|
|
2EX1
 
 | | Crystal structure of mutifunctional sialyltransferase from Pasteurella multocida with CMP bound | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, a2,3-sialyltransferase, a2,a6-sialyltransferase | | Authors: | Ni, L, Sun, M, Chen, X, Fisher, A.J. | | Deposit date: | 2005-11-07 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytidine 5'-Monophosphate (CMP)-Induced Structural Changes in a Multifunctional Sialyltransferase from Pasteurella multocida
Biochemistry, 45, 2006
|
|
7CY6
 
 | | Crystal Structure of CMD1 in complex with 5mC-DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*(5CM)P*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY7
 
 | | Crystal Structure of CMD1 in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*GP*CP*GP*CP*GP*GP*GP*A)-3'), FE (II) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY5
 
 | | Crystal Structure of CMD1 in complex with vitamin C | | Descriptor: | ASCORBIC ACID, CITRIC ACID, FE (III) ION, ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY8
 
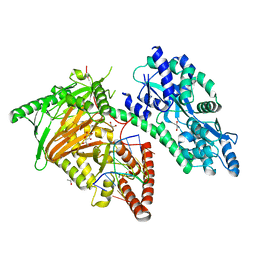 | | Crystal Structure of CMD1 in complex with 5mC-DNA and vitamin C | | Descriptor: | 1,2-ETHANEDIOL, ASCORBIC ACID, DNA (5'-D(P*(5CM)P*GP*CP*GP*CP*GP*GP*GP*A)-3'), ... | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
7CY4
 
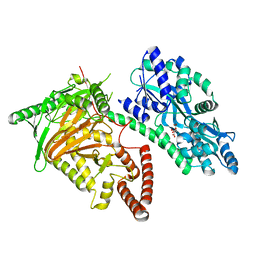 | | Crystal Structure of CMD1 in apo form | | Descriptor: | CITRIC ACID, FE (III) ION, Maltodextrin-binding protein,5-methylcytosine-modifying enzyme 1 | | Authors: | Li, W, Zhang, T, Sun, M, Ding, J. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism for vitamin C-derived C 5 -glyceryl-methylcytosine DNA modification catalyzed by algal TET homologue CMD1.
Nat Commun, 12, 2021
|
|
1T61
 
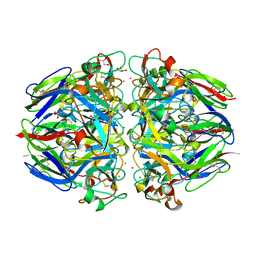 | | crystal structure of collagen IV NC1 domain from placenta basement membrane | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
1T60
 
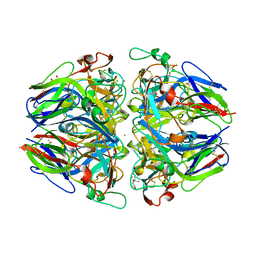 | | Crystal structure of Type IV collagen NC1 domain from bovine lens capsule | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
2I4I
 
 | | Crystal Structure of human DEAD-box RNA helicase DDX3X | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent RNA helicase DDX3X | | Authors: | Hogbom, M, Karlberg, T, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Johansson, I, Kotenyova, T, Magnusdottir, A, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Wallden, K, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-22 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Conserved Domains 1 and 2 of the Human DEAD-box Helicase DDX3X in Complex with the Mononucleotide AMP
J.Mol.Biol., 372, 2007
|
|
2IBN
 
 | | Crystal structure of Human myo-Inositol Oxygenase (MIOX) | | Descriptor: | (2S,3R,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXANONE, CYSTEINE, FE (III) ION, ... | | Authors: | Hallberg, B.M, Busam, R.D, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Johansson, I, Karlberg, T, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Sagemark, J, Stenmark, P, Sundstrom, M, Uppenberg, J, Van Den Berg, S, Weigelt, J, Thorsell, A.G, Persson, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-11 | | Release date: | 2006-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biophysical Characterization of Human myo-Inositol Oxygenase
J.Biol.Chem., 283, 2008
|
|
2I59
 
 | | Solution structure of RGS10 | | Descriptor: | Regulator of G-protein signaling 10 | | Authors: | Fedorov, O, Higman, V.A, Diehl, A, Leidert, M, Lemak, A, Schmieder, P, Oschkinat, H, Elkins, J, Soundarajan, M, Doyle, D.A, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-24 | | Release date: | 2006-10-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
138D
 
 | | A-DNA DECAMER D(GCGGGCCCGC)-HEXAGONAL CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
137D
 
 | | A-DNA DECAMER D(GCGGGCCCGC)-ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
2J2C
 
 | | Crystal structure of Human Cytosolic 5'-Nucleotidase II (NT5C2, cN-II) | | Descriptor: | CYTOSOLIC PURINE 5'-NUCLEOTIDASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Karlberg, T, Kotenyova, T, Loppnau, P, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2006-08-16 | | Release date: | 2006-09-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Human Cytosolic 5'- Nucleotidase II: Insights Into Allosteric Regulation and Substrate Recognition
J.Biol.Chem., 282, 2007
|
|
116D
 
 | |
117D
 
 | |
2J4E
 
 | | THE ITP COMPLEX OF HUMAN INOSINE TRIPHOSPHATASE | | Descriptor: | INOSINE 5'-TRIPHOSPHATE, INOSINE TRIPHOSPHATE PYROPHOSPHATASE, INOSINIC ACID, ... | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmbergschiavone, L, Hogbom, M, Kotenyova, T, Landry, R, Loppnau, P, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Schuler, H, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-08-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Inosine Triphosphatase: Substrate Binding and Implication of the Inosine Triphosphatase Deficiency Mutation P32T.
J.Biol.Chem., 282, 2007
|
|
