3AJJ
 
 | | Crystal Structure of d(CGCGGATf5UCGCG): 5-Formyluridine/Guanosine Base-pair in B-DNA | | 分子名称: | 5'-D(*CP*GP*CP*GP*GP*AP*TP*(UFR)P*CP*GP*CP*G)-3' | | 著者 | Tsunoda, M, Sakaue, T, Ueno, Y, Matsuda, A, Takenaka, A. | | 登録日 | 2010-06-07 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | Insights into the structures of DNA damaged by hydroxyl radical: crystal structures of DNA duplexes containing 5-formyluracil
J Nucleic Acids, 2010, 2010
|
|
2BPP
 
 | |
2CPO
 
 | | CHLOROPEROXIDASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLOROPEROXIDASE, ... | | 著者 | Sundaramoorthy, M, Poulos, T.L. | | 登録日 | 1996-02-10 | | 公開日 | 1997-02-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of chloroperoxidase: a heme peroxidase--cytochrome P450 functional hybrid.
Structure, 3, 1995
|
|
2DLC
 
 | | Crystal structure of the ternary complex of yeast tyrosyl-tRNA synthetase | | 分子名称: | MAGNESIUM ION, O-(ADENOSINE-5'-O-YL)-N-(L-TYROSYL)PHOSPHORAMIDATE, T-RNA (76-MER), ... | | 著者 | Tsunoda, M, Kusakabe, Y, Tanaka, N, Nakamura, K.T. | | 登録日 | 2006-04-18 | | 公開日 | 2007-06-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms.
Nucleic Acids Res., 35, 2007
|
|
3UD2
 
 | | Crystal structure of Selenomethionine ZU5A-ZU5B protein domains of human erythrocyte ankyrin | | 分子名称: | Ankyrin-1, CHLORIDE ION, ETHANOL, ... | | 著者 | Yasunaga, M, Ipsaro, J.J, Mondragon, A. | | 登録日 | 2011-10-27 | | 公開日 | 2012-02-22 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structurally Similar but Functionally Diverse ZU5 Domains in Human Erythrocyte Ankyrin.
J.Mol.Biol., 417, 2012
|
|
3NGL
 
 | | Crystal structure of bifunctional 5,10-methylenetetrahydrofolate dehydrogenase / cyclohydrolase from Thermoplasma acidophilum | | 分子名称: | Bifunctional protein folD, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Sung, M.W, Lee, W.H, Hwang, K.Y. | | 登録日 | 2010-06-12 | | 公開日 | 2011-04-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of bifunctional 5,10-methylenetetrahydrofolate dehydrogenase/cyclohydrolase from Thermoplasma acidophilum
Biochem.Biophys.Res.Commun., 406, 2011
|
|
2KFV
 
 | | Structure of the amino-terminal domain of human FK506-binding protein 3 / Northeast Structural Genomics Consortium Target HT99A | | 分子名称: | FK506-binding protein 3 | | 著者 | Sunnerhagen, M, Davis, T, Gutmanas, A, Fares, C, Ouyang, H, Lemak, A, Li, Y, Weigelt, J, Bountra, C, Edwards, A, Arrowsmith, C.H, Dhe-Paganon, S, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | 登録日 | 2009-02-27 | | 公開日 | 2009-06-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the N-terminal domain of FK506-binding protein 3
To be Published
|
|
2ETZ
 
 | | The NMR minimized average structure of the Itk SH2 domain bound to a phosphopeptide | | 分子名称: | Lymphocyte cytosolic protein 2 phosphopeptide fragment, Tyrosine-protein kinase ITK/TSK | | 著者 | Sundd, M, Pletneva, E.V, Fulton, D.B, Andreotti, A.H. | | 登録日 | 2005-10-27 | | 公開日 | 2006-02-07 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Details of Itk Activation by Prolyl Isomerization and Phospholigand Binding: The NMR Structure of the Itk SH2 Domain Bound to a Phosphopeptide.
J.Mol.Biol., 357, 2006
|
|
2EU0
 
 | | The NMR ensemble structure of the Itk SH2 domain bound to a phosphopeptide | | 分子名称: | Lymphocyte cytosolic protein 2 phosphopeptide fragment, Tyrosine-protein kinase ITK/TSK | | 著者 | Sundd, M, Pletneva, E.V, Fulton, D.B, Andreotti, A.H. | | 登録日 | 2005-10-27 | | 公開日 | 2006-02-07 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Details of Itk Activation by Prolyl Isomerization and Phospholigand Binding: The NMR Structure of the Itk SH2 Domain Bound to a Phosphopeptide.
J.Mol.Biol., 357, 2006
|
|
5N88
 
 | | Crystal structure of antibody bound to viral protein | | 分子名称: | PC4 and SFRS1-interacting protein, VH59 antibody | | 著者 | Bao, L, Hannon, C, Cruz-Migoni, A, Ptchelkine, D, Sun, M.-y, Derveni, M, Bunjobpol, W, Chambers, J.S, Simmons, A, Phillips, S.E.V, Rabbitts, T.H. | | 登録日 | 2017-02-23 | | 公開日 | 2017-12-20 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Intracellular immunization against HIV infection with an intracellular antibody that mimics HIV integrase binding to the cellular LEDGF protein.
Sci Rep, 7, 2017
|
|
6XE0
 
 | | Cryo-EM structure of NusG-CTD bound to 70S ribosome (30S: NusG-CTD fragment) | | 分子名称: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Washburn, R, Zuber, P, Sun, M, Hashem, Y, Shen, B, Li, W, Harvey, S, Acosta-Reyes, F.J, Knauer, S.H, Frank, J, Gottesman, M.E. | | 登録日 | 2020-06-11 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | Escherichia coli NusG Links the Lead Ribosome with the Transcription Elongation Complex.
Iscience, 23, 2020
|
|
2RCY
 
 | | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound | | 分子名称: | GLYCEROL, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Wernimont, A.K, Lew, J, Lin, Y.H, Ren, H, Sun, X, Khuu, C, Hassanali, A, Wasney, G, Zhao, Y, Kozieradzki, I, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | 登録日 | 2007-09-20 | | 公開日 | 2007-10-23 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of Plasmodium falciparum pyrroline carboxylate reductase (MAL13P1.284) with NADP bound.
To be Published
|
|
6KSR
 
 | |
2R77
 
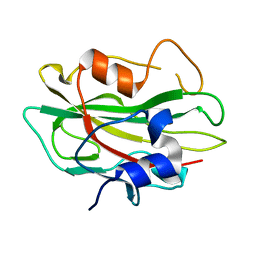 | | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum | | 分子名称: | Phosphatidylethanolamine-binding protein, putative | | 著者 | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Crombette, L, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | 登録日 | 2007-09-07 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum.
To be Published
|
|
2RHD
 
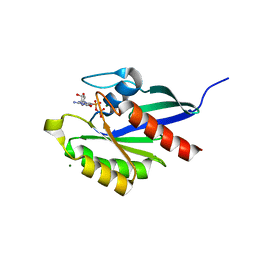 | | Crystal structure of Cryptosporidium parvum small GTPase RAB1A | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Small GTP binding protein rab1a | | 著者 | Dong, A, Xu, X, Lew, J, Lin, Y.H, Khuu, C, Sun, X, Qiu, W, Kozieradzki, I, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Sukumar, D, Structural Genomics Consortium (SGC) | | 登録日 | 2007-10-09 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Crystal structure of Cryptosporidium parvum small GTPase RAB1A.
To be Published
|
|
6ORL
 
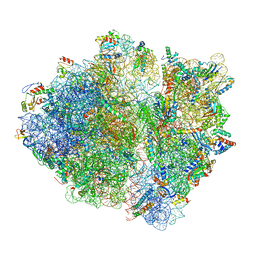 | | RF1 pre-accommodated 70S complex at 24 ms | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-04-30 | | 公開日 | 2019-06-19 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OT3
 
 | | RF2 accommodated state bound Release complex 70S at 24 ms | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-05-02 | | 公開日 | 2019-06-19 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OST
 
 | | RF2 pre-accommodated state bound Release complex 70S at 24ms | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-05-02 | | 公開日 | 2019-06-19 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSK
 
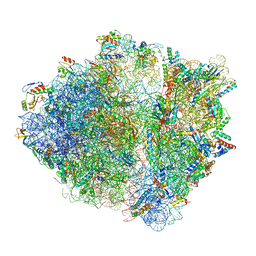 | | RF1 accommodated 70S complex at 60 ms | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-05-01 | | 公開日 | 2019-06-26 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OSQ
 
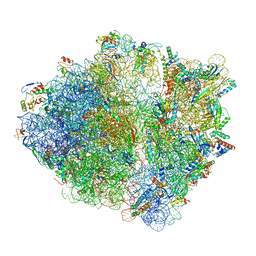 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-05-02 | | 公開日 | 2019-06-26 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | 分子名称: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | 著者 | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-11 | | 最終更新日 | 2021-04-21 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
6OUO
 
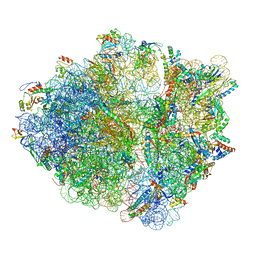 | | RF2 accommodated state bound 70S complex at long incubation time | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | 登録日 | 2019-05-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-11 | | 最終更新日 | 2021-04-21 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
409D
 
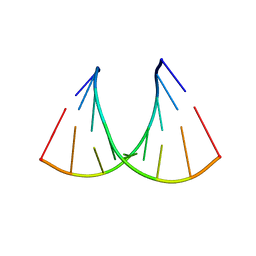 | | CRYSTAL STRUCTURE OF AN RNA R(CCCIUGGG) WITH THREE INDEPENDENT DUPLEXES INCORPORATING TANDEM I.U WOBBLES | | 分子名称: | RNA (5'-R(*CP*CP*CP*IP*UP*GP*GP*G)-3') | | 著者 | Pan, B, Mitra, S.N, Sun, L, Hart, D, Sundaralingam, M. | | 登録日 | 1998-06-26 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of an RNA octamer duplex r(CCCIUGGG)2 incorporating tandem I.U wobbles.
Nucleic Acids Res., 26, 1998
|
|
