6DCE
 
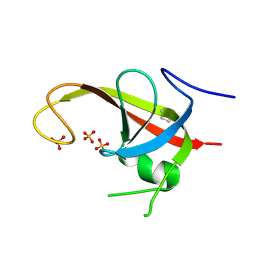 | | X-ray structure of FIP200 claw domain | | Descriptor: | RB1-inducible coiled-coil protein 1, SULFATE ION | | Authors: | Su, M.-Y, Hurley, J.H. | | Deposit date: | 2018-05-05 | | Release date: | 2019-03-06 | | Last modified: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates.
Mol. Cell, 74, 2019
|
|
8W4J
 
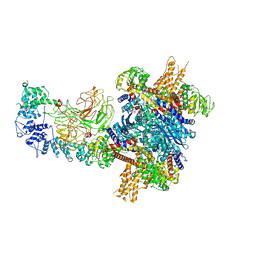 | |
5K7B
 
 | | Beclin 2 CCD homodimer | | Descriptor: | Beclin-2 | | Authors: | Su, M, Sinha, S. | | Deposit date: | 2016-05-25 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | BECN2 interacts with ATG14 through a metastable coiled-coil to mediate autophagy.
Protein Sci., 26, 2017
|
|
8DMG
 
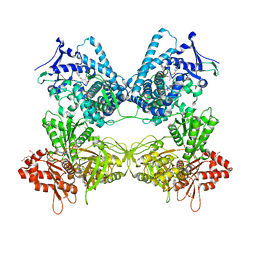 | | CYP102A1 in Closed Conformation | | Descriptor: | 6-methoxy-2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-1H-benzimidazole, Bifunctional cytochrome P450/NADPH--P450 reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Su, M, Xu, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Insight into the conformational dynamics of cytochrome P450 CYP102A1 enzyme using Cryo-EM
To Be Published
|
|
8DME
 
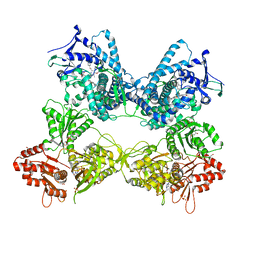 | | CYP102A1 in Open Conformation | | Descriptor: | 6-methoxy-2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-1H-benzimidazole, Bifunctional cytochrome P450/NADPH--P450 reductase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Su, M, Xu, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Insight into the conformational dynamics of cytochrome P450 CYP102A1 enzyme using Cryo-EM
To Be Published
|
|
7MGE
 
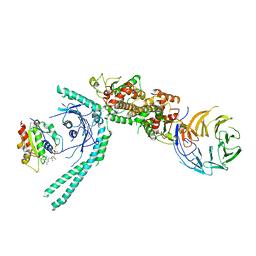 | | Structure of C9orf72:SMCR8:WDR41 in complex with ARF1 | | Descriptor: | ADP-ribosylation factor 1, BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Su, M.-Y, Hurley, J.H. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis for the ARF GAP activity and specificity of the C9orf72 complex.
Nat Commun, 12, 2021
|
|
4R8F
 
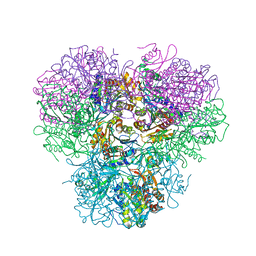 | |
5K9L
 
 | | Beclin 2 CCD N187L mutant homodimer | | Descriptor: | Beclin-2 | | Authors: | Su, M, Sinha, S. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | BECN2 interacts with ATG14 through a metastable coiled-coil to mediate autophagy.
Protein Sci., 26, 2017
|
|
2M5V
 
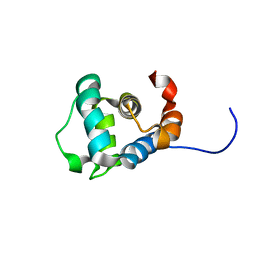 | |
8KHP
 
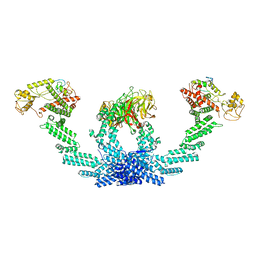 | | CULLIN3-KLHL22-RBX1 E3 ligase | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch-like protein 22 | | Authors: | Su, M.-Y, Su, M.-Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
5WQL
 
 | |
2N2G
 
 | |
2N3P
 
 | |
8THK
 
 | |
8THL
 
 | |
4XH6
 
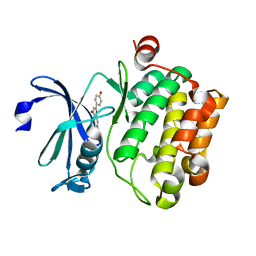 | | Crystal structure of proto-oncogene kinase Pim1 bound to hispidulin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-6-methoxy-4H-chromen-4-one, Serine/threonine-protein kinase pim-1 | | Authors: | Su, M.-Y, Chang, C.-I. | | Deposit date: | 2015-01-05 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Total Synthesis of Hispidulin and the Structural Basis for Its Inhibition of Proto-oncogene Kinase Pim-1
J.Nat.Prod., 78, 2015
|
|
8KGY
 
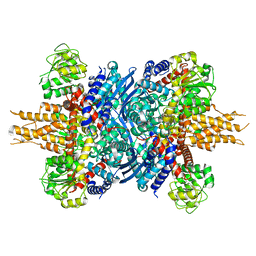 | | Human glutamate dehydrogenase I | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Su, M.-Y. | | Deposit date: | 2023-08-20 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
6V4U
 
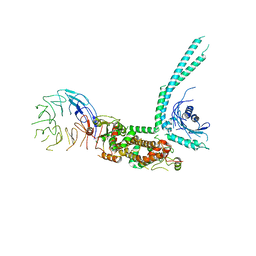 | | Cryo-EM structure of SMCR8-C9orf72-WDR41 complex | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Su, M.Y, Hurley, J.H. | | Deposit date: | 2019-12-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the C9orf72 ARF GAP complex that is haploinsufficient in ALS and FTD.
Nature, 585, 2020
|
|
8DCS
 
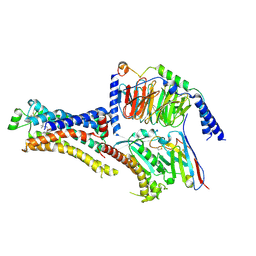 | | Cryo-EM structure of cyanopindolol-bound beta1-adrenergic receptor in complex with heterotrimeric Gs-protein | | Descriptor: | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, Endolysin,Endolysin,Beta-1 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Su, M, Paknejad, N, Hite, R.K, Huang, X.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of beta 1 -adrenergic receptor in complex with Gs and ligands of different efficacies.
Nat Commun, 13, 2022
|
|
8DCR
 
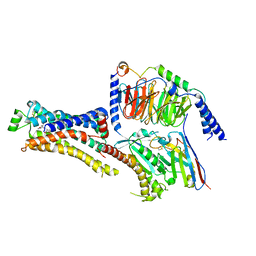 | | Cryo-EM structure of dobutamine-bound beta1-adrenergic receptor in complex with heterotrimeric Gs-protein | | Descriptor: | DOBUTAMINE, Endolysin,Endolysin,Beta-1 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Su, M, Paknejad, N, Hite, R.K, Huang, X.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of beta 1 -adrenergic receptor in complex with Gs and ligands of different efficacies.
Nat Commun, 13, 2022
|
|
4MI8
 
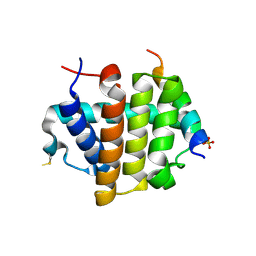 | |
6B9X
 
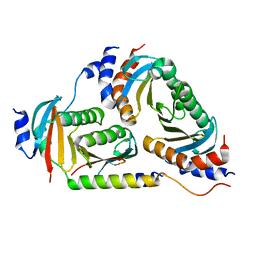 | | Crystal structure of Ragulator | | Descriptor: | Hepatitis B virus x interacting protein, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | Authors: | SU, M.-Y, Hurley, J.H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex.
Mol. Cell, 68, 2017
|
|
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
5WUC
 
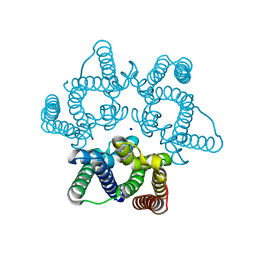 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SODIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUD
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
