4YQM
 
 | | Glutathione S-transferase Omega 1 bound to covalent inhibitor C1-27 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-N-[4-chloro-3-(dimethylsulfamoyl)phenyl]acetamide, Glutathione S-transferase omega-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor.
Nat Commun, 7, 2016
|
|
4YQV
 
 | | Glutathione S-transferase Omega 1 bound to covalent inhibitor C4-10 | | Descriptor: | 2-(ethylsulfanyl)-N-methyl-N-[(1-phenyl-1H-pyrazol-4-yl)methyl]acetamide, Glutathione S-transferase omega-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor.
Nat Commun, 7, 2016
|
|
4YQU
 
 | | Glutathione S-transferase Omega 1 bound to covalent inhibitor C1-31 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase omega-1, N-{5-(azepan-1-ylsulfonyl)-2-[(ethylsulfanyl)methoxy]phenyl}acetamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor.
Nat Commun, 7, 2016
|
|
4G8O
 
 | | Crystal Structure of a novel small molecule inactivator bound to plasminogen activator inhibitor-1 | | Descriptor: | (2S)-3-({[3-(trifluoromethyl)phenoxy]carbonyl}amino)propane-1,2-diyl bis(3,4,5-trihydroxybenzoate), 1,2-ETHANEDIOL, Plasminogen activator inhibitor 1, ... | | Authors: | Stuckey, J.A, Lawrence, D.A, Li, S.-H. | | Deposit date: | 2012-07-23 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanistic characterization and crystal structure of a small molecule inactivator bound to plasminogen activator inhibitor-1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4G8R
 
 | | Crystal Structure of a novel small molecule inactivator bound to plasminogen activator inhibitor-1 | | Descriptor: | (2S)-3-({[3-(trifluoromethyl)phenoxy]carbonyl}amino)propane-1,2-diyl bis(3,4,5-trihydroxybenzoate), Plasminogen activator inhibitor-1, SULFATE ION | | Authors: | Stuckey, J.A, Lawrence, D.A, Li, S.-H. | | Deposit date: | 2012-07-23 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mechanistic characterization and crystal structure of a small molecule inactivator bound to plasminogen activator inhibitor-1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3LQ6
 
 | |
8SX4
 
 | | Crystal Structure of eIF4e in complex with Compound 7n | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E, [(~{Z})-4-[2-azanyl-7-[(5-chloranyl-1~{H}-indol-2-yl)methyl]-6-oxidanylidene-1~{H}-purin-9-yl]but-2-enyl]phosphonic acid | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Design of Cell-Permeable Inhibitors of Eukaryotic Translation Initiation Factor 4E (eIF4E) for Inhibiting Aberrant Cap-Dependent Translation in Cancer.
J.Med.Chem., 66, 2023
|
|
2R8Z
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli in complex with a phosphate and a calcium ion | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CALCIUM ION, PHOSPHATE ION | | Authors: | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | Deposit date: | 2007-09-11 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
2R8X
 
 | | Crystal structure of YrbI phosphatase from Escherichia coli | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CHLORIDE ION | | Authors: | Tsodikov, O.V, Aggarwal, P, Rubin, J.R, Stuckey, J.A, Woodard, R.W, Biswas, T. | | Deposit date: | 2007-09-11 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Tail of KdsC: CONFORMATIONAL CHANGES CONTROL THE ACTIVITY OF A HALOACID DEHALOGENASE SUPERFAMILY PHOSPHATASE.
J.Biol.Chem., 284, 2009
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
3TWE
 
 | | Crystal Structure of the de novo designed peptide alpha4H | | Descriptor: | ACETYL GROUP, TRIETHYLENE GLYCOL, alpha4H | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TWG
 
 | |
3TWF
 
 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3a | | Descriptor: | ACETYL GROUP, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7UC7
 
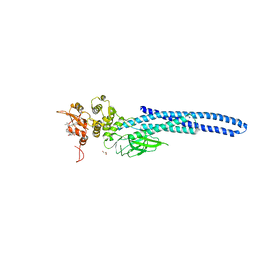 | | Stat5a Core in complex with Compound 17 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(1-benzofuran-5-yl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7UBT
 
 | | Stat5a Core in complex with Compound 18 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(1,3-benzothiazol-5-yl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7UC6
 
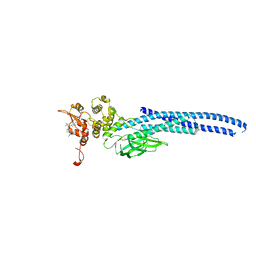 | | Stat5a Core in complex with Compound 12 | | Descriptor: | N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(4-bromophenyl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7TVB
 
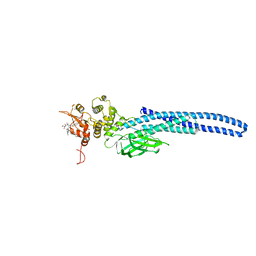 | | Stat5A Core in Complex with AK305 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N,N-dimethyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7TVA
 
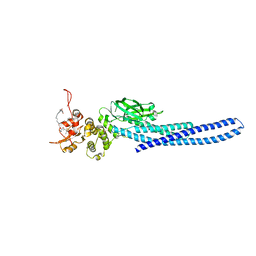 | | Stat5a Core in complex with AK2292 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MALONATE ION, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N-(5-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}pent-4-yn-1-yl)-N-methyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.835 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
6EGP
 
 | |
4TW3
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
5JFE
 
 | | Flavin-dependent thymidylate synthase with H2-dUMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-uridylic acid, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sapra, A, Stuckey, J, Palfey, B. | | Deposit date: | 2016-04-19 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Evaluating H2-dUMP as an Intermediate in the oxidation of Flavin-dependent Thymidylate Synthase
To Be Published
|
|
6W7G
 
 | | Structure of EED bound to inhibitor 1056 | | Descriptor: | 8-(2,6-dimethylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, FORMIC ACID, Polycomb protein EED, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
6W7F
 
 | | Structure of EED bound to inhibitor 5285 | | Descriptor: | 8-(6-cyclopropylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, GLYCEROL, Polycomb protein EED | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
4J09
 
 | | Crystal Structure of LpxA bound to RJPXD33 | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, ... | | Authors: | Jenkins, R.J, Meagher, J.L, Stuckey, J.A, Dotson, G.D. | | Deposit date: | 2013-01-30 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Recognition of Peptide RJPXD33 by Acyltransferases in Lipid A Biosynthesis.
J.Biol.Chem., 289, 2014
|
|
7MSB
 
 | | Structure of EED bound to EEDi-4259 | | Descriptor: | (9aM,12aS)-12-{[(5-fluoro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,11,12a-tetraazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
