3HD6
 
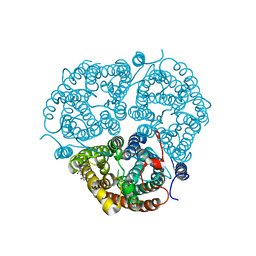 | | Crystal Structure of the Human Rhesus Glycoprotein RhCG | | Descriptor: | Ammonium transporter Rh type C, octyl beta-D-glucopyranoside | | Authors: | Gruswitz, F, Chaudhary, S, Ho, J.D, Pezeshki, B, Ho, C.-M, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-05-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Function of human Rh based on structure of RhCG at 2.1 A.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3K8N
 
 | |
2TSC
 
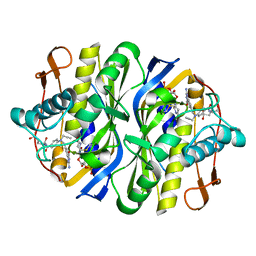 | | STRUCTURE, MULTIPLE SITE BINDING, AND SEGMENTAL ACCOMODATION IN THYMIDYLATE SYNTHASE ON BINDING D/UMP AND AN ANTI-FOLATE | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Montfort, W.R, Stroud, R.M. | | Deposit date: | 1991-07-03 | | Release date: | 1991-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure, multiple site binding, and segmental accommodation in thymidylate synthase on binding dUMP and an anti-folate.
Biochemistry, 29, 1990
|
|
2TGD
 
 | |
3R1F
 
 | | Crystal structure of a key regulator of virulence in Mycobacterium tuberculosis | | Descriptor: | ESX-1 secretion-associated regulator EspR | | Authors: | Rosenberg, O.S, Dovey, C, Finer-Moore, J, Stroud, R.M, Cox, J.S. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-03 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | EspR, a key regulator of Mycobacterium tuberculosis virulence, adopts a unique dimeric structure among helix-turn-helix proteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2NG1
 
 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Freymann, D.M, Stroud, R.M, Walter, P. | | Deposit date: | 1998-09-11 | | Release date: | 1999-07-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP.
Nat.Struct.Biol., 6, 1999
|
|
4GGM
 
 | | Structure of LpxI | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, MAGNESIUM ION, UDP-2,3-diacylglucosamine pyrophosphatase LpxI | | Authors: | Metzger IV, L.E, Lee, J.K, Finer-Moore, J.S, Raetz, C.R.H, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-03 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | LpxI structures reveal how a lipid A precursor is synthesized.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4HB1
 
 | | A DESIGNED FOUR HELIX BUNDLE PROTEIN. | | Descriptor: | DHP1, UNKNOWN ATOM OR ION | | Authors: | Schafmeister, C.E, Laporte, S.L, Miercke, L.J.W, Stroud, R.M. | | Deposit date: | 1997-11-10 | | Release date: | 1998-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A designed four helix bundle protein with native-like structure.
Nat.Struct.Biol., 4, 1997
|
|
4ISK
 
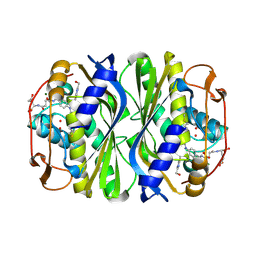 | | Crystal structure of E.coli thymidylate synthase with dUMP and the BGC 945 inhibitor | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2'-deoxy-5'-uridylic acid, MAGNESIUM ION, ... | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2013-01-16 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Development and Binding Mode Assessment of N-[4-[2-Propyn-1-yl[(6S)-4,6,7,8-tetrahydro-2-(hydroxymethyl)-4-oxo-3H-cyclopenta[g]quinazolin-6-yl]amino]benzoyl]-l-gamma-glutamyl-d-glutamic Acid (BGC 945), a Novel Thymidylate Synthase Inhibitor That Targets Tumor Cells.
J.Med.Chem., 56, 2013
|
|
4ITS
 
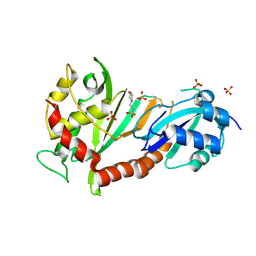 | | Crystal structure of the catalytic domain of human Pus1 with MES in the active site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, tRNA pseudouridine synthase A, ... | | Authors: | Czudnochowski, N, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2013-01-18 | | Release date: | 2013-06-05 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Human Pseudouridine Synthase 1 (hPus1), a C-Terminal Helical Insert Blocks tRNA from Binding in the Same Orientation as in the Pus1 Bacterial Homologue TruA, Consistent with Their Different Target Selectivities.
J.Mol.Biol., 425, 2013
|
|
1TGN
 
 | |
4ZYR
 
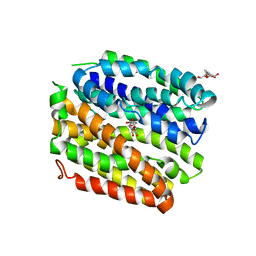 | | Crystal structure of E. coli Lactose permease G46W/G262W bound to p-nitrophenyl alpha-D-galactopyranoside (alpha-NPG) | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, nonyl beta-D-glucopyranoside | | Authors: | Kumar, H, Finer-Moore, J.S, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | Structure of LacY with an alpha-substituted galactoside: Connecting the binding site to the protonation site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1NG1
 
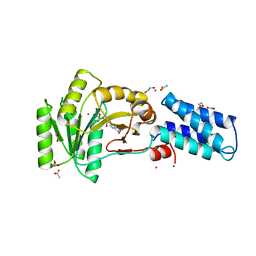 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CADMIUM ION, ... | | Authors: | Freymann, D.M, Stroud, R.M, Walter, P. | | Deposit date: | 1998-04-30 | | Release date: | 1999-07-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP.
Nat.Struct.Biol., 6, 1999
|
|
3LNN
 
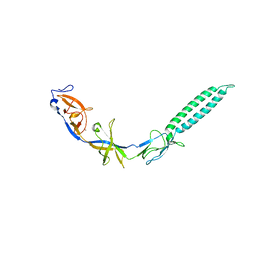 | | Crystal structure of ZneB from Cupriavidus metallidurans | | Descriptor: | Membrane fusion protein (MFP) heavy metal cation efflux ZneB (CzcB-like), ZINC ION | | Authors: | Lee, J.K, De Angelis, F, Miercke, L.J, Stroud, R.M, Vandenbussche, G, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-02-02 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2IST
 
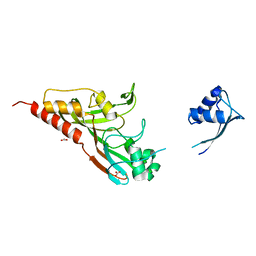 | |
3NK5
 
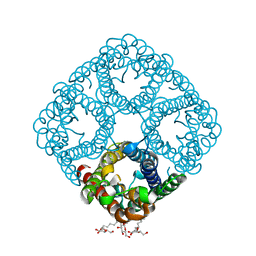 | | Crystal structure of AqpZ mutant F43W | | Descriptor: | Aquaporin Z, octyl beta-D-glucopyranoside | | Authors: | Savage, D.F, O'Connell, J.D, Stroud, R.M, Finer-Moore, J.S. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural context shapes the aquaporin selectivity filter.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MP7
 
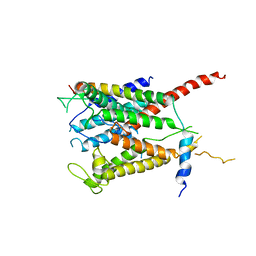 | |
1NN6
 
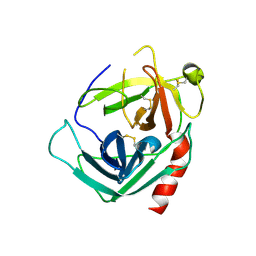 | | Human Pro-Chymase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase | | Authors: | Reiling, K.K, Krucinski, J, Miercke, L.J.W, Raymond, W.W, Caughey, G.H, Stroud, R.M. | | Deposit date: | 2003-01-12 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human pro-chymase: a model for the activating transition of granule-associated proteases.
Biochemistry, 42, 2003
|
|
3NKC
 
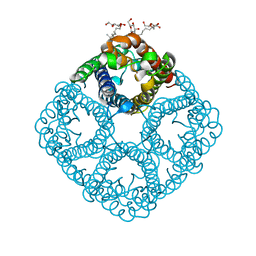 | | Crystal structure of AqpZ F43W,H174G,T183F | | Descriptor: | Aquaporin Z, octyl beta-D-glucopyranoside | | Authors: | Savage, D.F, O'Connell III, J.D, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural context shapes the aquaporin selectivity filter.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NE2
 
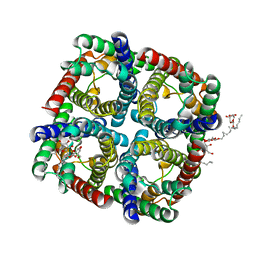 | |
3NKA
 
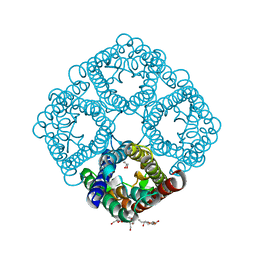 | | Crystal structure of AqpZ H174G,T183F | | Descriptor: | Aquaporin Z, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Savage, D.F, O'Connell III, J.D, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural context shapes the aquaporin selectivity filter.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3P5S
 
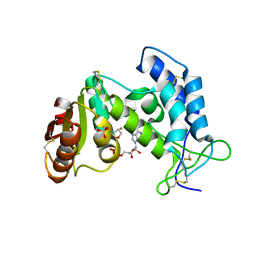 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD38 molecule, SULFATE ION, ... | | Authors: | Egea, P.F, Muller-Stauffler, H, Kohn, I, Cakou-Kefir, C, Stroud, R.M, Kellenberburger, E, Schuber, F. | | Deposit date: | 2010-10-10 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
1ZPR
 
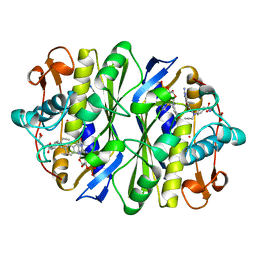 | | E. COLI THYMIDYLATE SYNTHASE MUTANT E58Q IN COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sage, C.R, Stout, T.J, Rutenber, E.E, Stroud, R.M. | | Deposit date: | 1996-10-15 | | Release date: | 1997-07-07 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An essential role for water in an enzyme reaction mechanism: the crystal structure of the thymidylate synthase mutant E58Q.
Biochemistry, 35, 1996
|
|
2G86
 
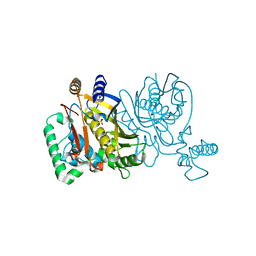 | |
2G8D
 
 | |
