6XVI
 
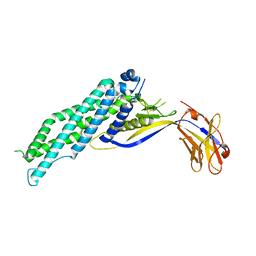 | |
6XUX
 
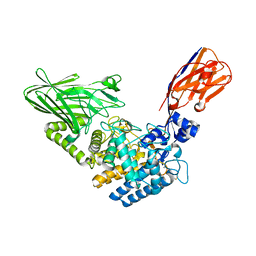 | | Crystal structure of Megabody Mb-Nb207-cYgjK_NO | | Descriptor: | CALCIUM ION, Nanobody,Glucosidase YgjK,Glucosidase YgjK,Nanobody | | Authors: | Steyaert, J, Uchanski, T, Fischer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.90000641 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
6XV8
 
 | |
8G8W
 
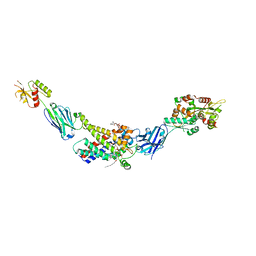 | | Molecular mechanism of nucleotide inhibition of human uncoupling protein 1 | | Descriptor: | CARDIOLIPIN, GUANOSINE-5'-TRIPHOSPHATE, Mitochondrial brown fat uncoupling protein 1, ... | | Authors: | Gogoi, P, Jones, S.A, Ruprecht, J.J, King, M.S, Lee, Y, Zogg, T, Pardon, E, Chand, D, Steimle, S, Copeman, D, Cotrim, C.A, Steyaert, J, Crichton, P.G, Moiseenkova-Bell, V, Kunji, E.R.S. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of purine nucleotide inhibition of human uncoupling protein 1.
Sci Adv, 9, 2023
|
|
3K74
 
 | | Disruption of protein dynamics by an allosteric effector antibody | | Descriptor: | Dihydrofolate reductase, Nanobody | | Authors: | Oyen, D, Srinivasan, V, Steyaert, J, Barlow, J. | | Deposit date: | 2009-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Constraining enzyme conformational change by an antibody leads to hyperbolic inhibition.
J.Mol.Biol., 407, 2011
|
|
2X1P
 
 | | Gelsolin Nanobody | | Descriptor: | GELSOLIN NANOBODY | | Authors: | Van Den Abbeele, A, Declercq, S, De Ganck, A, De Corte, V, Van Loo, B, Srinivasan, V, Steyaert, J, Van De Kerckhove, J, Gettemans, J. | | Deposit date: | 2010-01-03 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Llama-Derived Gelsolin Single-Domain Antibody Blocks Gelsolin-G-Actin Interaction.
Cell.Mol.Life Sci., 67, 2010
|
|
2X89
 
 | | Structure of the Beta2_microglobulin involved in amyloidogenesis | | Descriptor: | ANTIBODY, BETA-2-MICROGLOBULIN | | Authors: | Domanska, K, Srinivasan, V, Vanderhaegen, S, Pardon, E, Marquez, J.A, Bellotti, V, Wyns, L, Steyaert, J. | | Deposit date: | 2010-03-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Atomic Structure of a Nanobody-Trapped Domain-Swapped Dimer of an Amyloidogenic {Beta}2-Microglobulin Variant.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1YOE
 
 | |
4MQS
 
 | | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Muscarinic acetylcholine receptor M2, Nanobody 9-8 | | Authors: | Kruse, A.C, Ring, A.M, Manglik, A, Hu, J, Hu, K, Eitel, K, Huebner, H, Pardon, E, Valant, C, Sexton, P.M, Christopoulos, A, Felder, C.C, Gmeiner, P, Steyaert, J, Weis, W.I, Garcia, K.C, Wess, J, Kobilka, B.K. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature, 504, 2013
|
|
8V8K
 
 | | Crystal Structure of Nanobody NbE | | Descriptor: | Nanobody NbE | | Authors: | Koehl, A, Manglik, A, Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Steyaert, J, Ballet, S, Boland, A, Stoeber, M. | | Deposit date: | 2023-12-05 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of mu-opioid receptor targeting by a nanobody antagonist.
Nat Commun, 15, 2024
|
|
4MQT
 
 | | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo and allosteric modulator LY2119620 | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Muscarinic acetylcholine receptor M2, ... | | Authors: | Kruse, A.C, Ring, A.M, Manglik, A, Hu, J, Hu, K, Eitel, K, Huebner, H, Pardon, E, Valant, C, Sexton, P.M, Christopoulos, A, Felder, C.C, Gmeiner, P, Steyaert, J, Weis, W.I, Garcia, K.C, Wess, J, Kobilka, B.K. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature, 504, 2013
|
|
4N9O
 
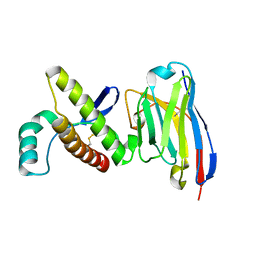 | | Probing the N-terminal beta-sheet conversion in the crystal structure of the human prion protein bound to a Nanobody | | Descriptor: | Major prion protein, Nanobody Nb484 | | Authors: | Abskharon, R.N.N, Giachin, G, Wohlkonig, A, Soror, S.H, Pardon, E, Legname, G, Steyaert, J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the N-Terminal beta-Sheet Conversion in the Crystal Structure of the Human Prion Protein Bound to a Nanobody.
J.Am.Chem.Soc., 136, 2014
|
|
6I53
 
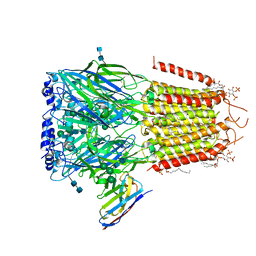 | | Cryo-EM structure of the human synaptic alpha1-beta3-gamma2 GABAA receptor in complex with Megabody38 in a lipid nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Laverty, D, Desai, R, Uchanski, T, Masiulis, S, Wojciech, J.S, Malinauskas, T, Zivanov, J, Pardon, E, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human alpha 1 beta 3 gamma 2 GABAAreceptor in a lipid bilayer.
Nature, 565, 2019
|
|
1HYF
 
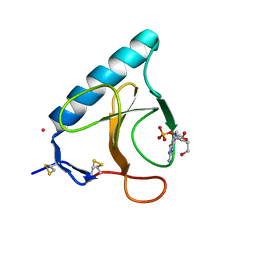 | | RIBONUCLEASE T1 V16A MUTANT IN COMPLEX WITH SR2+ | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, GUANYL-SPECIFIC RIBONUCLEASE T1, STRONTIUM ION | | Authors: | De Swarte, J, De Vos, S, Langhorst, U, Steyaert, J, Loris, R. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The contribution of metal ions to the conformational stability of ribonuclease T1: crystal versus solution.
Eur.J.Biochem., 268, 2001
|
|
1HZ1
 
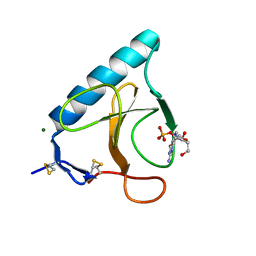 | | RIBONUCLEASE T1 V16A MUTANT IN COMPLEX WITH MG2+ | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, MAGNESIUM ION, RIBONUCLEASE T1 | | Authors: | De Swarte, J, De Vos, S, Langhorst, U, Steyaert, J, Loris, R. | | Deposit date: | 2001-01-23 | | Release date: | 2001-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The contribution of metal ions to the conformational stability of ribonuclease T1: crystal versus solution.
Eur.J.Biochem., 268, 2001
|
|
1I0V
 
 | | Ribonuclease T1 in complex with 2'GMP (form I crystal) | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | De Swarte, J, De Vos, S, Langhorst, U, Steyaert, J, Loris, R. | | Deposit date: | 2001-01-30 | | Release date: | 2001-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.234 Å) | | Cite: | The contribution of metal ions to the conformational stability of ribonuclease T1: crystal versus solution.
Eur.J.Biochem., 268, 2001
|
|
1I0X
 
 | | RIBONUCLEASE T1 IN COMPLEX WITH 2'GMP (FORM II CRYSTAL) | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | De Swarte, J, De Vos, S, Langhorst, U, Steyaert, J, Loris, R. | | Deposit date: | 2001-01-30 | | Release date: | 2001-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The contribution of metal ions to the conformational stability of ribonuclease T1: crystal versus solution.
Eur.J.Biochem., 268, 2001
|
|
1HP0
 
 | | CRYSTAL STRUCTURE OF AN INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE FROM TRYPANOSOMA VIVAX IN COMPLEX WITH THE SUBSTRATE ANALOGUE 3-DEAZA-ADENOSINE | | Descriptor: | 3-DEAZA-ADENOSINE, CALCIUM ION, INOSINE-ADENOSINE-GUANOSINE-PREFERRING NUCLEOSIDE HYDROLASE | | Authors: | Versees, W, Decanniere, K, Pelle, R, Depoorter, J, Parkin, D.W, Steyaert, J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of a novel purine specific nucleoside hydrolase from Trypanosoma vivax.
J.Mol.Biol., 307, 2001
|
|
1I2E
 
 | | Ribonuclease T1 V16A mutant, form I | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | De Vos, S, Backmann, J, Steyaert, J, Loris, R. | | Deposit date: | 2001-02-09 | | Release date: | 2001-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrophobic core manipulations in ribonuclease T1
Biochemistry, 40, 2001
|
|
1BIR
 
 | | RIBONUCLEASE T1, PHE 100 TO ALA MUTANT COMPLEXED WITH 2' GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Doumen, J, Gonciarz, M, Zegers, I, Loris, R, Wyns, L, Steyaert, J. | | Deposit date: | 1996-01-04 | | Release date: | 1996-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A catalytic function for the structurally conserved residue Phe 100 of ribonuclease T1.
Protein Sci., 5, 1996
|
|
7B5G
 
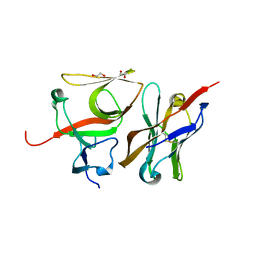 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS3(Nb14527) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody Nb14527, ... | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2020-12-03 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
6B73
 
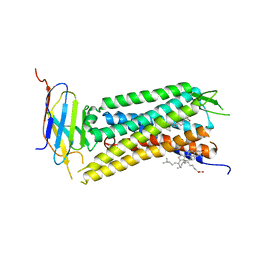 | | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor | | Descriptor: | CHOLESTEROL, N-[(5alpha,6beta)-17-(cyclopropylmethyl)-3-hydroxy-7,8-didehydro-4,5-epoxymorphinan-6-yl]-3-iodobenzamide, Nanobody, ... | | Authors: | Che, T, Majumdar, S, Zaidi, S.A, Kormos, C, McCorvy, J.D, Wang, S, Mosier, P.D, Uprety, R, Vardy, E, Krumm, B.E, Han, G.W, Lee, M.Y, Pardon, E, Steyaert, J, Huang, X.P, Strachan, R.T, Tribo, A.R, Pasternak, G.W, Carroll, I.F, Stevens, R.C, Cherezov, V, Katritch, V, Wacker, D, Roth, B.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor.
Cell, 172, 2018
|
|
6C9W
 
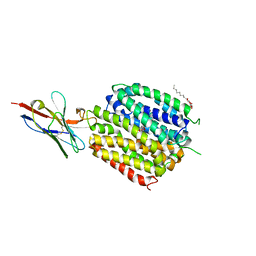 | | Crystal Structure of a ligand bound LacY/Nanobody Complex | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, Nanobody9047, ... | | Authors: | Kumar, H, Finer-Moore, J.S, Jiang, X, Smirnova, I, Kasho, V, Pardon, E, Steyaert, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a ligand-bound LacY-Nanobody Complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ZRA
 
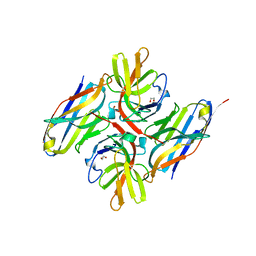 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS1(Nb14497) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody NbSOS1 (Nb14497) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2022-05-04 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
6EQI
 
 | | Structure of PINK1 bound to ubiquitin | | Descriptor: | GLYCEROL, Nb696, Serine/threonine-protein kinase PINK1, ... | | Authors: | Schubert, A.F, Gladkova, C, Pardon, E, Wagstaff, J.L, Freund, S.M.V, Steyaert, J, Maslen, S, Komander, D. | | Deposit date: | 2017-10-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of PINK1 in complex with its substrate ubiquitin.
Nature, 552, 2017
|
|
