6C0J
 
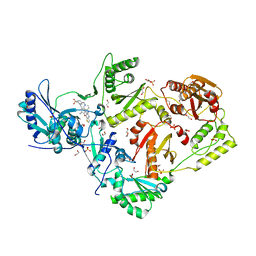 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
4Z3S
 
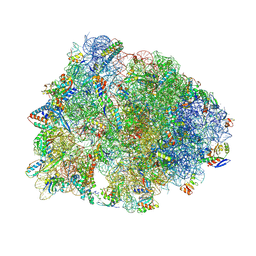 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic A201A, mRNA and three tRNAs in the A, P and E sites at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 3'-deoxy-3'-{[(2E)-3-(4-{[(4Z)-6-O-(6-deoxy-3,4-di-O-methyl-alpha-D-mannopyranosyl)-5-O-methyl-alpha-D-threo-hex-4-enofuranosyl]oxy}phenyl)-2-methylprop-2-enoyl]amino}-N,N-dimethyladenosine, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-03-31 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
6C0R
 
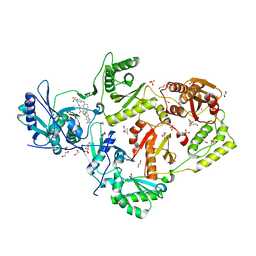 | | Crystal structure of HIV-1 K103N/Y181C mutant reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6C0K
 
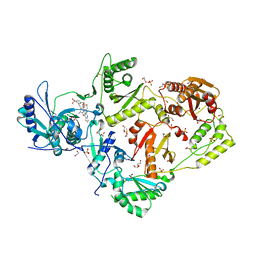 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6CGF
 
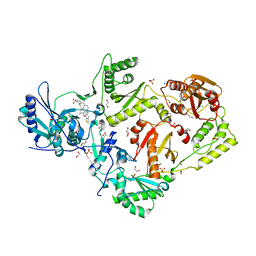 | | Crystal structure of HIV-1 Y188L mutant reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-02-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6C0O
 
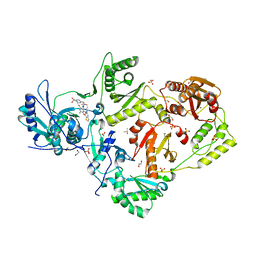 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6C0L
 
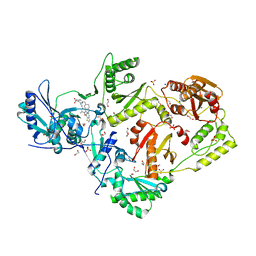 | | Crystal structure of HIV-1 E138K mutant reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
4WQY
 
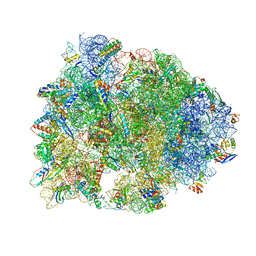 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G in the post-translocational state (without fusitic acid) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
4Z8C
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome bound to translation inhibitor oncocin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Roy, R.N, Lomakin, I.B, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2015-04-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4Y4O
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome with rRNA modifications and bound to protein Y (YfiA) at 2.3A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Polikanov, Y.S, Melnikov, S.V, Soll, D, Steitz, T.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1QU3
 
 | | INSIGHTS INTO EDITING FROM AN ILE-TRNA SYNTHETASE STRUCTURE WITH TRNA(ILE) AND MUPIROCIN | | Descriptor: | ISOLEUCYL-TRNA, ISOLEUCYL-TRNA SYNTHETASE, MUPIROCIN, ... | | Authors: | Silvian, L.F, Wang, J, Steitz, T.A. | | Deposit date: | 1999-07-06 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin.
Science, 285, 1999
|
|
2REB
 
 | |
3I56
 
 | | Co-crystal structure of Triacetyloleandomcyin Bound to the Large Ribosomal Subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Steitz, T.A, Moore, P.B. | | Deposit date: | 2009-07-03 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of triacetyloleandomycin and mycalamide A bind to the large ribosomal subunit of Haloarcula marismortui.
Antimicrob.Agents Chemother., 53, 2009
|
|
1FFK
 
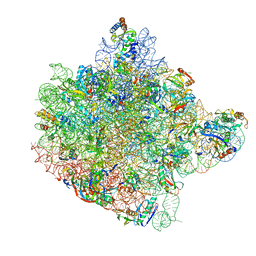 | | CRYSTAL STRUCTURE OF THE LARGE RIBOSOMAL SUBUNIT FROM HALOARCULA MARISMORTUI AT 2.4 ANGSTROM RESOLUTION | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Ban, N, Nissen, P, Hansen, J, Moore, P.B, Steitz, T.A. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The complete atomic structure of the large ribosomal subunit at 2.4 A resolution.
Science, 289, 2000
|
|
3CFR
 
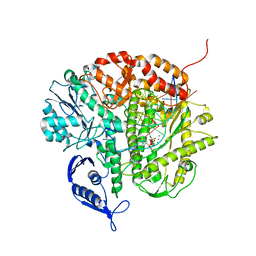 | | Structure of the replicating complex of a POL Alpha family DNA Polymerase, ternary complex 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DGP*DGP*DAP*DCP*DTP*DGP*DCP*DTP*DTP*DAP*(DOC))-3'), ... | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
1QU2
 
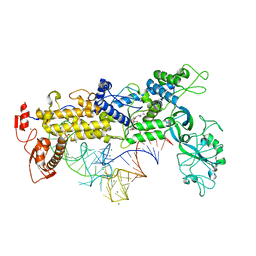 | | INSIGHTS INTO EDITING FROM AN ILE-TRNA SYNTHETASE STRUCTURE WITH TRNA(ILE) AND MUPIROCIN | | Descriptor: | ISOLEUCYL-TRNA, ISOLEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, ... | | Authors: | Silvian, L.F, Wang, J, Steitz, T.A. | | Deposit date: | 1999-07-06 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin.
Science, 285, 1999
|
|
4M6F
 
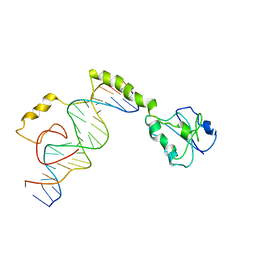 | |
3G71
 
 | | Co-crystal structure of Bruceantin bound to the large ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome.
J.Mol.Biol., 389, 2009
|
|
3OVB
 
 | | How the CCA-adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CCA-Adding Enzyme, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
3G4S
 
 | | Co-crystal structure of Tiamulin bound to the large ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome.
J.Mol.Biol., 389, 2009
|
|
3G6E
 
 | | Co-crystal structure of Homoharringtonine bound to the large ribosomal subunit | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 23S ribosomal RNA, 50S ribosomal protein L10E, ... | | Authors: | Gurel, G, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome.
J.Mol.Biol., 389, 2009
|
|
3I55
 
 | | Co-crystal structure of Mycalamide A Bound to the Large Ribosomal Subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Steitz, T.A, Moore, P.B. | | Deposit date: | 2009-07-03 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structures of triacetyloleandomycin and mycalamide A bind to the large ribosomal subunit of Haloarcula marismortui.
Antimicrob.Agents Chemother., 53, 2009
|
|
3OVS
 
 | | How the CCA-adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CCA-Adding Enzyme, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
3HL2
 
 | | The crystal structure of the human SepSecS-tRNASec complex | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Monothiophosphate, O-phosphoseryl-tRNA(Sec) selenium transferase, ... | | Authors: | Palioura, S, Steitz, T.A, Soll, D, Simonovic, M. | | Deposit date: | 2009-05-26 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation.
Science, 325, 2009
|
|
2HPI
 
 | | Eubacterial and Eukaryotic Replicative DNA Polymerases are not Homologous: X-ray Structure of DNA Polymerase III | | Descriptor: | CHLORIDE ION, DNA polymerase III alpha subunit, MAGNESIUM ION, ... | | Authors: | Bailey, S, Wing, R.A, Steitz, T.A. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of T. aquaticus DNA Polymerase III Is Distinct from Eukaryotic Replicative DNA Polymerases.
Cell(Cambridge,Mass.), 126, 2006
|
|
