5ACR
 
 | | W228Y-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | CALCIUM ION, GIM-1 PROTEIN, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
2Y87
 
 | | Native VIM-7. Structural and computational investigations of VIM-7: Insights into the substrate specificity of VIM metallo-beta- lactamases | | Descriptor: | MAGNESIUM ION, METALLO-B-LACTAMASE, UNKNOWN ATOM OR ION, ... | | Authors: | Saradhi, P, Leiros, H.-K.S, Ahmad, R, Spencer, J, Leiros, I, Walsh, T.R, Sundsfjord, A, Samuelsen, O. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Computational Investigations of Vim- 7: Insights Into the Substrate Specificity of Vim Metallo-Beta-Lactamases
J.Mol.Biol., 411, 2011
|
|
5ACP
 
 | | W228R-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | GIM-1 PROTEIN, MAGNESIUM ION, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
5ACT
 
 | | W228S-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | GIM-1 PROTEIN, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
5ACQ
 
 | | W228A-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | BETA-LACTAMASE, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
5ACS
 
 | | Y233A-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | GIM-1 PROTEIN, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
4EQI
 
 | | Crystal structure of serratia fonticola carbapenemase SFC-1 | | Descriptor: | 1,2-ETHANEDIOL, Carbapenem-hydrolizing beta-lactamase SFC-1, SODIUM ION | | Authors: | Fonseca, F, Spencer, J. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The basis for carbapenem hydrolysis by class A beta-lactamases: a combined investigation using crystallography and simulations.
J.Am.Chem.Soc., 134, 2012
|
|
4EUZ
 
 | | Crystal structure of serratia fonticola carbapenemase SFC-1 S70A-Meropenem complex | | Descriptor: | (4R,5S,6S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-6-[(1R)-1-hydroxyethyl]-4-methyl-7-oxo-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, Carbapenem-hydrolizing beta-lactamase SFC-1, ... | | Authors: | Fonseca, F, Spencer, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The basis for carbapenem hydrolysis by class A beta-lactamases: a combined investigation using crystallography and simulations.
J.Am.Chem.Soc., 134, 2012
|
|
6ZYP
 
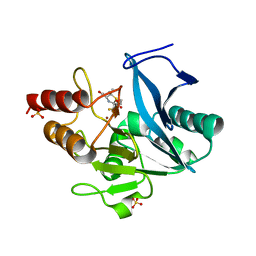 | | Structure of NDM-1 with 2-Mercaptomethyl-thiazolidine L-anti-1b | | Descriptor: | (2~{S},4~{R})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
6ZYQ
 
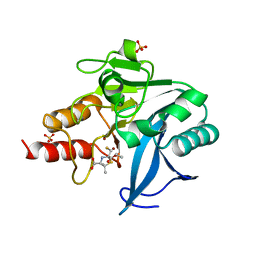 | | Structure of NDM-1 with 2-Mercaptomethyl-thiazolidine D-syn-1b | | Descriptor: | (2~{S},4~{S})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
2J66
 
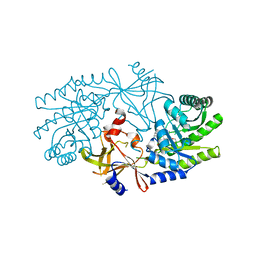 | | Structural characterisation of BtrK decarboxylase from butirosin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, BTRK, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Popovic, B, Li, Y, Chirgadze, D.Y, Blundell, T.L, Spencer, J.B. | | Deposit date: | 2006-09-26 | | Release date: | 2006-09-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterisation of Btrk Decarboxylase from Bacillus Circulans Butirosin Biosynthesis
To be Published
|
|
6ZYO
 
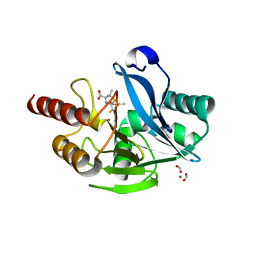 | | Structure of VIM-2 with 2-Mercaptomethyl-thiazolidine D-syn-1b | | Descriptor: | (2~{S},4~{S})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
6ZYR
 
 | | Structure of IMP-1 with 2-Mercaptomethyl-thiazolidine L-anti-1b | | Descriptor: | (2~{S},4~{R})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
6ZYS
 
 | | Structure of IMP-1 with 2-Mercaptomethyl-thiazolidine D-syn-1b | | Descriptor: | (2~{S},4~{S})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase IMP-1, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87000954 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
6ZYN
 
 | | Structure of VIM-2 with 2-Mercaptomethyl-thiazolidine L-anti-1b | | Descriptor: | (2~{S},4~{R})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4000138 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
7BH4
 
 | | XFEL structure of apo CTX-M-15 after mixing for 0.7 sec with ertapenem using a piezoelectric injector (PolyPico) | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BH5
 
 | | XFEL structure of the ertapenem-derived CTX-M-15 acylenzyme after mixing for 2 sec using a piezoelectric injector (PolyPico) | | Descriptor: | (2~{S},3~{R},4~{R})-3-[5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-4-methyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BH7
 
 | | Room temperature, serial X-ray structure of the ertapenem-derived acylenzyme of CTX-M-15 (10 min soak) collected on fixed target chips at Diamond Light Source I24 | | Descriptor: | (2~{S},3~{R},4~{R})-3-[5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-4-methyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BH6
 
 | | Room temperature, serial X-ray structure of CTX-M-15 collected on fixed target chips at Diamond Light Source I24 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BH3
 
 | | XFEL structure of CTX-M-15 resting state | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
3H4T
 
 | | Chimeric Glycosyltransferase for the generation of novel natural products - GtfAH1 in complex with UDP-2F-Glc | | Descriptor: | Glycosyltransferase GtfA, Glycosyltransferase, PHOSPHATE ION, ... | | Authors: | Dias, M.V.B, Truman, A.W, Wu, S, Blundell, T.L, Huang, F, Spencer, J.B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Chimeric glycosyltransferases for the generation of hybrid glycopeptides
Chem.Biol., 16, 2009
|
|
3H4I
 
 | | Chimeric Glycosyltransferase for the generation of novel natural products | | Descriptor: | Glycosyltransferase GtfA, Glycosyltransferase, SULFATE ION, ... | | Authors: | Dias, M.V.B, Truman, A.W, Wu, S, Blundell, T.L, Huang, F, Spencer, J.B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Chimeric glycosyltransferases for the generation of hybrid glycopeptides
Chem.Biol., 16, 2009
|
|
4EV4
 
 | | Crystal structure of serratia fonticola carbapenemase SFC-1 E166A mutant with the acylenzyme intermediate of meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Carbapenem-hydrolizing beta-lactamase SFC-1 | | Authors: | Fonseca, F, Spencer, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The basis for carbapenem hydrolysis by class A beta-lactamases: a combined investigation using crystallography and simulations.
J.Am.Chem.Soc., 134, 2012
|
|
1GHX
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GHY
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(3-HYDROXY-PYRIDIN-2-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
