6U8W
 
 | | Crystal structure of DNMT3B(K777A)-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Zhang, Z.M, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.94891548 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6UX2
 
 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6AGO
 
 | | Crystal structure of MRG15-ASH1L Histone methyltransferase complex | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, Mortality factor 4 like 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Lee, Y, Song, J. | | Deposit date: | 2018-08-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis of MRG15-Mediated Activation of the ASH1L Histone Methyltransferase by Releasing an Autoinhibitory Loop.
Structure, 27, 2019
|
|
8T12
 
 | |
8T13
 
 | |
1G11
 
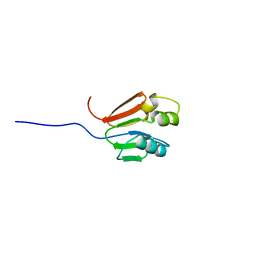 | | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR PROTEIN NMR STRUCTURE | | Descriptor: | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR | | Authors: | Hemmi, H, Studts, J.M, Chae, Y.K, Song, J, Markley, J.L, Fox, B.G. | | Deposit date: | 2000-10-10 | | Release date: | 2001-05-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the toluene 4-monooxygenase effector protein (T4moD).
Biochemistry, 40, 2001
|
|
1G10
 
 | | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR PROTEIN NMR STRUCTURE | | Descriptor: | TOLUENE-4-MONOOXYGENASE CATALYTIC EFFECTOR | | Authors: | Hemmi, H, Studts, J.M, Chae, Y.K, Song, J, Markley, J.L, Fox, B.G. | | Deposit date: | 2000-10-10 | | Release date: | 2001-05-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the toluene 4-monooxygenase effector protein (T4moD).
Biochemistry, 40, 2001
|
|
6PZV
 
 | |
8FZB
 
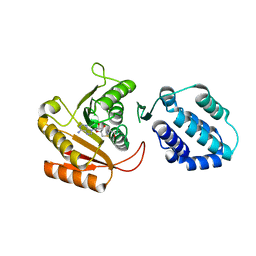 | | Crystal structure of human FAM86A | | Descriptor: | Protein-lysine N-methyltransferase EEF2KMT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shao, Z, Lu, J, Song, J. | | Deposit date: | 2023-01-28 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The FAM86 domain of FAM86A confers substrate specificity to promote EEF2-Lys525 methylation.
J.Biol.Chem., 299, 2023
|
|
6W8J
 
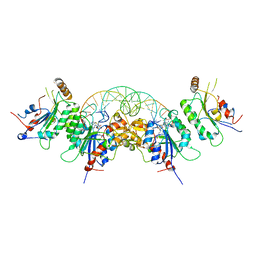 | | Structure of DNMT3A (R882H) in complex with CAG DNA | | Descriptor: | CAG DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
6W8D
 
 | | Structure of DNMT3A (R882H) in complex with CGT DNA | | Descriptor: | CGT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
6W8W
 
 | | Crystal structure of mouse DNMT1 in complex with CCG DNA | | Descriptor: | CCG DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), CCG DNA (5'-D(*CP*CP*TP*TP*CP*(C49)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
6W89
 
 | | Structure of DNMT3A (R882H) in complex with CGA DNA | | Descriptor: | CGA DNA (25-MER), CITRIC ACID, DNA (cytosine-5)-methyltransferase 3-like, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
6W8V
 
 | | Crystal structure of mouse DNMT1 in complex with ACG DNA | | Descriptor: | ACG DNA (5'-D(*AP*CP*TP*TP*AP*(C49)P*GP*GP*AP*AP*GP*G)-3'), ACG DNA (5'-D(*CP*CP*TP*TP*CP*(5CM)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
6W8B
 
 | | Structure of DNMT3A in complex with CGA DNA | | Descriptor: | CGA DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
6U8X
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpApG DNA | | Descriptor: | CpApG DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95063877 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
7L4C
 
 | | Crystal structure of the DRM2-CTT DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*TP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4K
 
 | | Crystal structure of the DRM2-CCG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4H
 
 | | Crystal structure of the DRM2-CTG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*TP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*AP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4N
 
 | | Crystal structure of the DRM2 (C397R)-CCG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4M
 
 | | Crystal structure of the DRM2-CCT DNA complex | | Descriptor: | DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*CP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4F
 
 | | Crystal structure of the DRM2-CAT DNA complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*AP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*TP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
8JBJ
 
 | | Crystal structure of anti-PVRIG Fab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Li, X.L, Song, J. | | Deposit date: | 2023-05-09 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Identification and characterization of an unexpected isomerization motif in CDRH2 that affects antibody activity.
Mabs, 15, 2023
|
|
3IKK
 
 | | Crystal structure analysis of msp domain | | Descriptor: | Vesicle-associated membrane protein-associated protein B/C | | Authors: | Shi, J, Lua, S, Song, J. | | Deposit date: | 2009-08-06 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elimination of the native structure and solubility of the hVAPB MSP domain by the Pro56Ser mutation that causes amyotrophic lateral sclerosis.
Biochemistry, 49, 2010
|
|
3FHQ
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | Jie, Y, Li, L, Shaw, N, Li, Y, Song, J, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.-C, Wang, L.-X, Wang, P, Liu, Z.-J. | | Deposit date: | 2008-12-10 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A
Plos One, 4, 2009
|
|
