5E6D
 
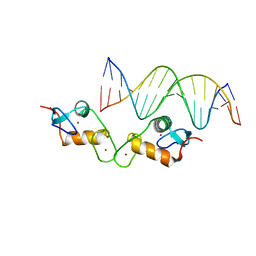 | | Glucocorticoid receptor DNA binding domain - ICAM1 NF-kB response element complex | | Descriptor: | DNA (5'-D(*GP*CP*TP*CP*CP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*GP*GP*AP*GP*C)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Rye, E.A, Herbst, A.G, Ortlund, E.A. | | Deposit date: | 2015-10-09 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cryptic glucocorticoid receptor-binding sites pervade genomic NF-kappa B response elements.
Nat Commun, 9, 2018
|
|
4MCE
 
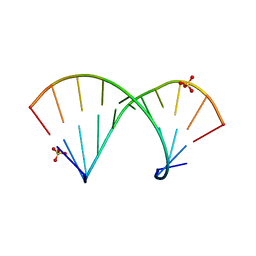 | | Crystal structure of the Gas5 GRE Mimic | | Descriptor: | Gas5 GREM Fwd, Gas5 GREM Rev, SULFATE ION | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2013-08-21 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate.
Nat Commun, 5, 2014
|
|
4MCF
 
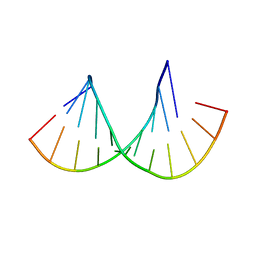 | | Crystal structure of the Gas5 GRE Mimic | | Descriptor: | Gas5 GREM Fwd, Gas5 GREM Rev, SULFATE ION | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2013-08-21 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate.
Nat Commun, 5, 2014
|
|
4TNT
 
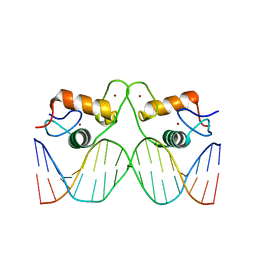 | | Structure of the human mineralocorticoid receptor in complex with DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), Mineralocorticoid receptor, ... | | Authors: | Hudson, W.H, Ortlund, E.A. | | Deposit date: | 2014-06-04 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3938 Å) | | Cite: | Crystal structure of the mineralocorticoid receptor DNA binding domain in complex with DNA.
Plos One, 9, 2014
|
|
1HRB
 
 | |
7DCL
 
 | |
7C7Z
 
 | |
2FTM
 
 | | Crystal structure of trypsin complexed with the BPTI variant (Tyr35->Gly) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Hanson, W.M, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rigidification of a Flexible Protease Inhibitor Variant upon Binding to Trypsin.
J.Mol.Biol., 366, 2007
|
|
2FTL
 
 | | Crystal structure of trypsin complexed with BPTI at 100K | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Hanson, W.M, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rigidification of a Flexible Protease Inhibitor Variant upon Binding to Trypsin.
J.Mol.Biol., 366, 2007
|
|
1KZF
 
 | | Crystal Structure of the Acyl-homoserine Lactone Synthase, EsaI | | Descriptor: | acyl-homoserinelactone synthase EsaI | | Authors: | Watson, W.T, Minogue, T.D, Val, D.L, Beck von Bodman, S, Churchill, M.E.A. | | Deposit date: | 2002-02-06 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing.
Mol.Cell, 9, 2002
|
|
4HN6
 
 | | GR DNA Binding Domain R460D/D462R - TSLP nGRE Complex | | Descriptor: | DNA (5'-D(*AP*GP*CP*TP*CP*TP*CP*CP*CP*GP*GP*AP*GP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*CP*TP*CP*CP*GP*GP*GP*AP*GP*AP*GP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Youn, C.E, Ortlund, E.A. | | Deposit date: | 2012-10-18 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | The structural basis of direct glucocorticoid-mediated transrepression.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4HN5
 
 | | GR DNA Binding Domain - TSLP nGRE Complex | | Descriptor: | DNA (5'-D(*AP*GP*CP*TP*CP*TP*CP*CP*CP*GP*GP*AP*GP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*CP*TP*CP*CP*GP*GP*GP*AP*GP*AP*GP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Youn, C.E, Ortlund, E.A. | | Deposit date: | 2012-10-18 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | The structural basis of direct glucocorticoid-mediated transrepression.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1APX
 
 | |
1CRN
 
 | |
1MLD
 
 | | REFINED STRUCTURE OF MITOCHONDRIAL MALATE DEHYDROGENASE FROM PORCINE HEART AND THE CONSENSUS STRUCTURE FOR DICARBOXYLIC ACID OXIDOREDUCTASES | | Descriptor: | CITRIC ACID, MALATE DEHYDROGENASE | | Authors: | Gleason, W.B, Fu, Z, Birktoft, J.J, Banaszak, L.J. | | Deposit date: | 1994-01-24 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined crystal structure of mitochondrial malate dehydrogenase from porcine heart and the consensus structure for dicarboxylic acid oxidoreductases.
Biochemistry, 33, 1994
|
|
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
4EWQ
 
 | | Human p38 alpha MAPK in complex with a pyridazine based inhibitor | | Descriptor: | 3-phenyl-4-(pyridin-4-yl)-6-[4-(pyrimidin-2-yl)piperazin-1-yl]pyridazine, 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, ACETATE ION, ... | | Authors: | Grum-Tokars, V, Minasov, G, Roy, S, Schavocky, J, Winsor, J, Watterson, D.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-27 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Selective and Brain Penetrant p38 alpha MAPK Inhibitor Candidate for Neurologic and Neuropsychiatric Disorders That Attenuates Neuroinflammation and Cognitive Dysfunction.
J.Med.Chem., 62, 2019
|
|
1G9M
 
 | | HIV-1 HXBC2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Kwong, P.D, Wyatt, R, Majeed, S, Robinson, J, Sweet, R.W, Sodroski, J, Hendrickson, W.A. | | Deposit date: | 2000-11-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates.
Structure Fold.Des., 8, 2000
|
|
1G9N
 
 | | HIV-1 YU2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Kwong, P.D, Wyatt, R, Majeed, S, Robinson, J, Sweet, R.W, Sodroski, J, Hendrickson, W.A. | | Deposit date: | 2000-11-25 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates.
Structure Fold.Des., 8, 2000
|
|
1GC1
 
 | | HIV-1 GP120 CORE COMPLEXED WITH CD4 AND A NEUTRALIZING HUMAN ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, CD4, ... | | Authors: | Kwong, P.D, Wyatt, R, Robinson, J, Sweet, R.W, Sodroski, J, Hendrickson, W.A. | | Deposit date: | 1998-06-15 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody.
Nature, 393, 1998
|
|
8B16
 
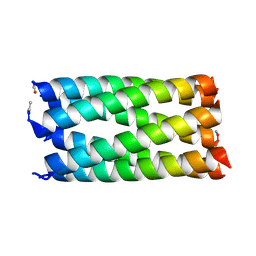 | |
4EGU
 
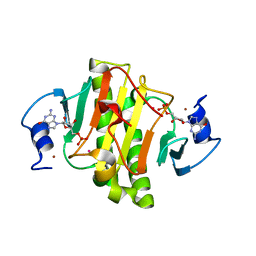 | | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION, ZINC ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-01 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile
To be Published
|
|
3GEU
 
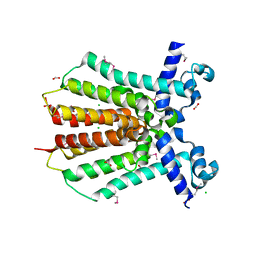 | | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family | | Descriptor: | CHLORIDE ION, FORMIC ACID, Intercellular adhesion protein R, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
To be Published
|
|
3FF1
 
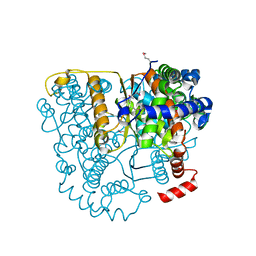 | | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glucose-6-phosphate isomerase, SODIUM ION | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Peterson, S, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus
TO BE PUBLISHED
|
|
5HM3
 
 | | 2.25 Angstrom Resolution Crystal Structure of Long-chain-fatty-acid-AMP Ligase FadD32 from Mycobacterium tuberculosis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Warwrzak, Z, Kuhn, M.L, Shuvalova, L, Flores, K.J, Wilson, D.J, Grimes, K.D, Aldrich, C.C, Anderson, W.A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-03 | | Last modified: | 2016-09-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Essential Mtb FadD32 Enzyme: A Promising Drug Target for Treating Tuberculosis.
Acs Infect Dis., 2, 2016
|
|
