7LSX
 
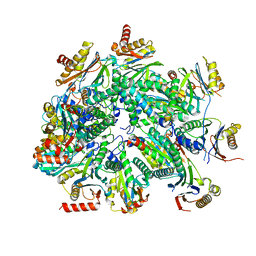 | | Cryo-EM structure of 13S proteasome core particle assembly intermediate purified from Pre3-1 proteasome mutant (G34D) | | Descriptor: | Proteasome assembly chaperone 2, Proteasome chaperone 1, Proteasome maturation factor UMP1, ... | | Authors: | Schnell, H.M, Walsh Jr, R.M, Rawson, S, Hanna, J.W. | | Deposit date: | 2021-02-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ULC
 
 | | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab at a nominal resolution of 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Chen, B, Harrison, S.C. | | Deposit date: | 2019-10-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16.
J.Mol.Biol., 432, 2020
|
|
3FC0
 
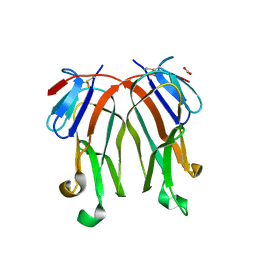 | | 1.8 A crystal structure of murine GITR ligand dimer expressed in Drosophila melanogaster S2 cells | | Descriptor: | ACETATE ION, GITR ligand | | Authors: | Chattopadhyay, K, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2008-11-20 | | Release date: | 2008-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.8 A structure of murine GITR ligand dimer expressed in Drosophila melanogaster S2 cells.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5AQI
 
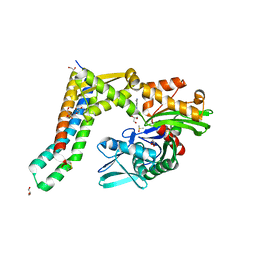 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | ADENINE, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, DIMETHYL SULFOXIDE, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AQQ
 
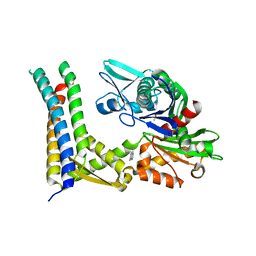 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7-methylquinazolin-4-amine, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AQR
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-methoxyquinazolin-4-amine, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
2ANO
 
 | | Crystal structure of E.coli dihydrofolate reductase in complex with NADPH and the inhibitor MS-SH08-17 | | Descriptor: | 1-{[N-(1-IMINO-GUANIDINO-METHYL)]SULFANYLMETHYL}-3-TRIFLUOROMETHYL-BENZENE, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
4DXD
 
 | | Staphylococcal Aureus FtsZ in complex with 723 | | Descriptor: | 3-[(6-chloro[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-difluorobenzamide, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Lu, J, Soisson, S.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Restoring methicillin-resistant Staphylococcus aureus susceptibility to beta-lactam antibiotics.
Sci Transl Med, 4, 2012
|
|
3NWM
 
 | | Crystal structure of a single chain construct composed of MHC class I H-2Kd, beta-2microglobulin and a peptide which is an autoantigen for type 1 diabetes | | Descriptor: | Peptide/beta-2microglobulin/MHC class I H-2Kd chimeric protein | | Authors: | Ramagopal, U.A, Samanta, D, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-07-09 | | Release date: | 2011-07-06 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of a single-chain peptide-MHC molecule that modulates both naive and activated CD8+ T cells.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7TKV
 
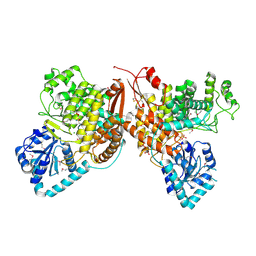 | | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Kim, Y, Crawford, M, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-17 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus
To Be Published
|
|
1DS3
 
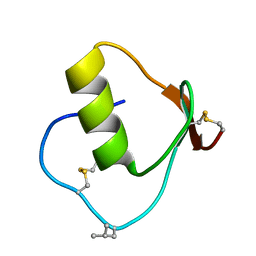 | | CRYSTAL STRUCTURE OF OMTKY3-CH2-ASP19I | | Descriptor: | OVOMUCOID | | Authors: | Bateman, K.S, Huang, K, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2000-01-06 | | Release date: | 2001-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
5GJE
 
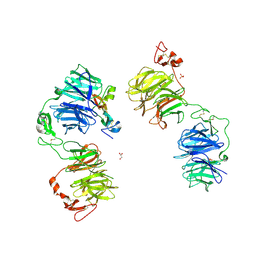 | | Three-dimensional reconstruction of human LRP6 ectodomain complexed with Dkk1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matoba, K, Mihara, E, Tamura-Kawakami, K, Hirai, H, Thompson, S, Iwasaki, K, Takagi, J. | | Deposit date: | 2016-06-29 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Conformational Freedom of the LRP6 Ectodomain Is Regulated by N-glycosylation and the Binding of the Wnt Antagonist Dkk1
Cell Rep, 18, 2017
|
|
3DA9
 
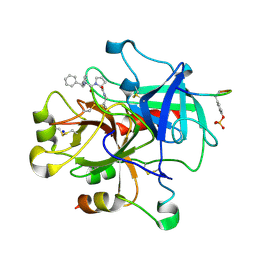 | |
1DK3
 
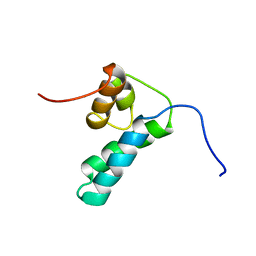 | | REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Maciejewski, M.W, Prasad, R, Liu, D.-J, Wilson, S.H, Mullen, G.P. | | Deposit date: | 1999-12-06 | | Release date: | 2000-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity.
J.Mol.Biol., 296, 2000
|
|
6UPH
 
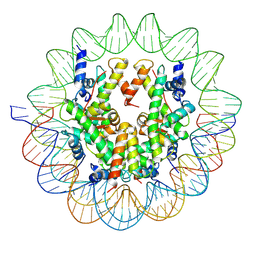 | | Structure of a Yeast Centromeric Nucleosome at 2.7 Angstrom resolution | | Descriptor: | DNA (119-MER), Histone H2A, Histone H2B.1, ... | | Authors: | Migl, D, Kschonsak, M, Arthur, C.P, Khin, Y, Harrison, S.C, Ciferri, C, Dimitrova, Y.N. | | Deposit date: | 2019-10-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryoelectron Microscopy Structure of a Yeast Centromeric Nucleosome at 2.7 angstrom Resolution.
Structure, 28, 2020
|
|
1DS2
 
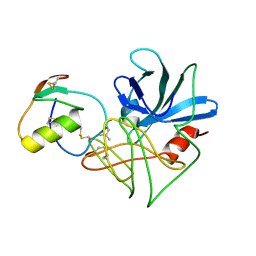 | | CRYSTAL STRUCTURE OF SGPB:OMTKY3-COO-LEU18I | | Descriptor: | OVOMUCOID, PROTEINASE B (SGPB) | | Authors: | Bateman, K.S, Huang, K, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2000-01-06 | | Release date: | 2001-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
8T0P
 
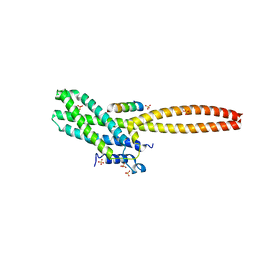 | | Structure of Cse4 bound to Ame1 and Okp1 | | Descriptor: | Histone H3-like centromeric protein CSE4, Inner kinetochore subunit AME1, Inner kinetochore subunit OKP1, ... | | Authors: | Deng, S, Harrison, S.C. | | Deposit date: | 2023-06-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Recognition of centromere-specific histone Cse4 by the inner kinetochore Okp1-Ame1 complex.
Embo Rep., 24, 2023
|
|
1DK2
 
 | | REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Maciejewski, M.W, Prasad, R, Liu, D.-J, Wilson, S.H, Mullen, G.P. | | Deposit date: | 1999-12-06 | | Release date: | 2000-02-14 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity.
J.Mol.Biol., 296, 2000
|
|
3FR4
 
 | | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte Fatty-Acid Binding Protein (A-FABP) inhibitors | | Descriptor: | 9-[2-(trifluoromethyl)benzyl]-2,3,4,9-tetrahydro-1H-carbazole-8-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Barf, T, Hammer, K, Lehmann, F, Haile, S, Axen, E, Medina, C, Uppenberg, J, Svensson, S, Rondahl, L, Lundb ck, T. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (A-FABP) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1DQT
 
 | | THE CRYSTAL STRUCTURE OF MURINE CTLA4 (CD152) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTOTOXIC T LYMPHOCYTE ASSOCIATED ANTIGEN 4 | | Authors: | Ostrov, D.A, Shi, W, Schwartz, J.C, Almo, S.C, Nathenson, S.G. | | Deposit date: | 2000-01-05 | | Release date: | 2000-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of murine CTLA-4 and its role in modulating T cell responsiveness.
Science, 290, 2000
|
|
3FR2
 
 | | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte Fatty-Acid Binding Protein (A-FABP) inhibitors | | Descriptor: | 9-benzyl-2,3,4,9-tetrahydro-1H-carbazole-8-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Barf, T, Hammer, K, Lehmann, F, Haile, S, Axen, E, Medina, C, Rondahl, L, Uppenberg, J, Svensson, S, Lundb ck, T. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (A-FABP) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2QQZ
 
 | | Crystal structure of putative glyoxalase family protein from Bacillus anthracis | | Descriptor: | GLYCEROL, Glyoxalase family protein, putative, ... | | Authors: | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Putative Glyoxalase Family Protein from Bacillus anthracis.
To be Published
|
|
7OXL
 
 | | Crystal structure of human Spermine Oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FAD-MDL72527 adduct, ... | | Authors: | Impagliazzo, A, Johannsson, S, Thomsen, M, Krapp, S. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human spermine oxidase in complex with a highly selective allosteric inhibitor.
Commun Biol, 5, 2022
|
|
3FR5
 
 | | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte Fatty-Acid Binding Protein (A-FABP) inhibitors | | Descriptor: | 5-(3-carbamoylbenzyl)-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-4-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Barf, T, Hammer, K, Lehmann, F, Haile, S, Axen, E, Medina, C, Uppenberg, J, Svensson, S, Rondahl, L, Lundb ck, T. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (A-FABP) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4Z72
 
 | | Crystal structure of inorganic pyrophosphatase from Mycobacterium tuberculosis in complex with two phosphate ions | | Descriptor: | CALCIUM ION, Inorganic pyrophosphatase, PHOSPHATE ION | | Authors: | Pratt, A.C, Biswas, T, Barnard-Britson, S, Tsodikov, O.V. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structural and computational dissection of the catalytic mechanism of the inorganic pyrophosphatase from Mycobacterium tuberculosis.
J.Struct.Biol., 192, 2015
|
|
