5ZG4
 
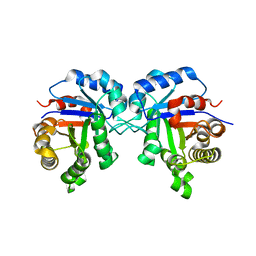 | | Crystal Structure of Triosephosphate isomerase SAD deletion mutant from Opisthorchis viverrini | | Descriptor: | Triosephosphate isomerase | | Authors: | Son, J, Kim, S, Kim, S.E, Lee, H, Lee, M.R, Hwang, K.Y. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Structural Analysis of an Epitope Candidate of Triosephosphate Isomerase in Opisthorchis viverrini.
Sci Rep, 8, 2018
|
|
2G2B
 
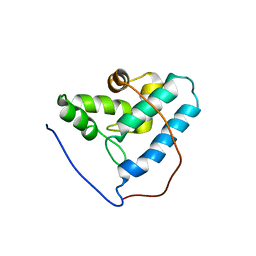 | | NMR structure of the human allograft inflammatory factor 1 | | Descriptor: | Allograft inflammatory factor 1 | | Authors: | Song, J, Tyler, R.C, Newman, C.L, Vinarov, D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-15 | | Release date: | 2006-02-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human allograft inflammatory factor 1
To be published
|
|
5HOO
 
 | |
4UQ6
 
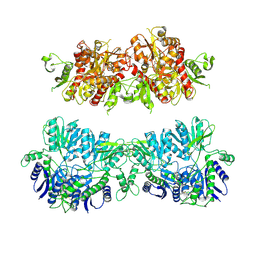 | | Electron density map of GluA2em in complex with LY451646 and glutamate | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQQ
 
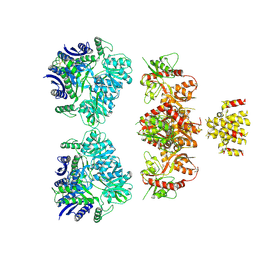 | | Electron density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, KAINATE 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQJ
 
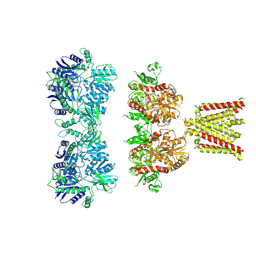 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | Descriptor: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQK
 
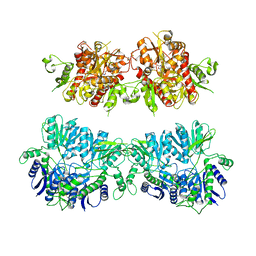 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
6E3J
 
 | |
6E3I
 
 | |
4ULL
 
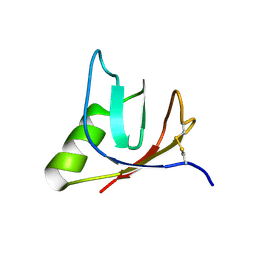 | | SOLUTION NMR STRUCTURE OF VEROTOXIN-1 B-SUBUNIT FROM E. COLI, 5 STRUCTURES | | Descriptor: | Shiga toxin 1B | | Authors: | Richardson, J.M, Evans, P.D, Homans, S.W, Donohue-Rolfe, A. | | Deposit date: | 1996-12-17 | | Release date: | 1997-04-01 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carbohydrate-binding B-subunit homopentamer of verotoxin VT-1 from E. coli.
Nat.Struct.Biol., 4, 1997
|
|
8W1N
 
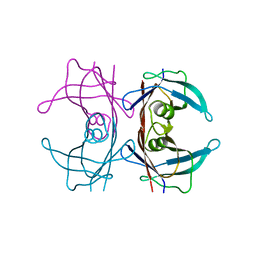 | |
8U8J
 
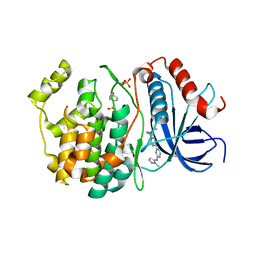 | |
8U8K
 
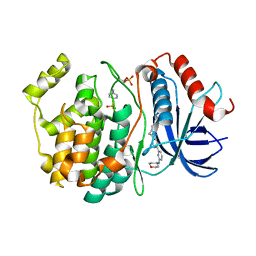 | |
8U1S
 
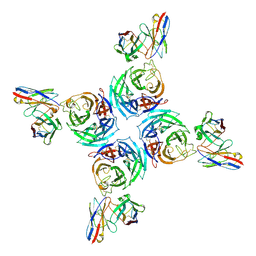 | |
8A4I
 
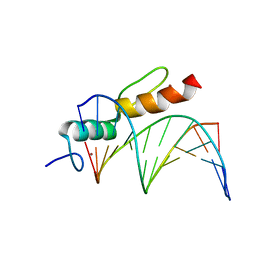 | | Crystal structure of SALL4 zinc finger cluster 4 with AT-rich DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*AP*TP*TP*AP*AP*TP*AP*TP*C)-3'), MAGNESIUM ION, Sal-like protein 4, ... | | Authors: | Watson, J.A, Pantier, R, Jayachandran, U, Chhatbar, K, Alexander-Howden, B, Kruusvee, V, Prendecki, M, Bird, A, Cook, A.G. | | Deposit date: | 2022-06-11 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure of SALL4 zinc finger domain reveals link between AT-rich DNA binding and Okihiro syndrome.
Life Sci Alliance, 6, 2023
|
|
5ML9
 
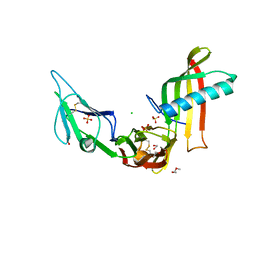 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer F4, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer F4 with specificity for Fc gamma receptor IIIa, CHLORIDE ION, ... | | Authors: | Robinson, J.I, Tomlinson, D.C, Baxter, E.W, Owen, R.L, Thomsen, M, Win, S.J, Nettleship, J.E, Tiede, C, Foster, R.J, Waterhouse, M.P, Harris, S.A, Owens, R.J, Fishwick, C.W.G, Goldman, A, McPherson, M.J, Morgan, A.W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MN2
 
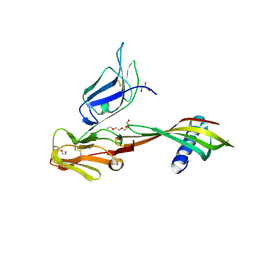 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer G3, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer G3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Robinson, J.I, Owen, R.L, Tomlinson, D.C, Baxter, E.W, Nettleship, J.E, Waterhouse, M.P, Harris, S.A, Owens, R.J, McPherson, M.J, Morgan, A.W, Tiede, C, Goldman, A, Thomsen, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8U1C
 
 | |
8WZU
 
 | |
8U1Q
 
 | |
8VCI
 
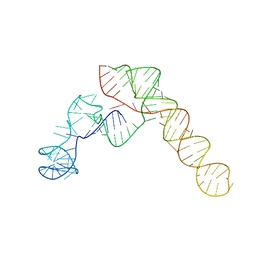 | | SARS-CoV-2 Frameshift Stimulatory Element with Upstream Multibranch Loop | | Descriptor: | Frameshift Stimulatory Element with Upstream Multi-branch Loop | | Authors: | Peterson, J.M, Becker, S.T, O'Leary, C.A, Juneja, P, Yang, Y, Moss, W.N. | | Deposit date: | 2023-12-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the SARS-CoV-2 Frameshift Stimulatory Element with an Upstream Multibranch Loop.
Biochemistry, 63, 2024
|
|
7UQ2
 
 | | Vs.4 from T4 phage in complex with cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CALCIUM ION, Vs.4 | | Authors: | Jenson, J.M, Chen, Z.J. | | Deposit date: | 2022-04-18 | | Release date: | 2023-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin-like conjugation by bacterial cGAS enhances anti-phage defence.
Nature, 616, 2023
|
|
6YMY
 
 | | Cytochrome c oxidase from Saccharomyces cerevisiae | | Descriptor: | (2R,5S,11R,14R)-5,8,11-trihydroxy-2-(nonanoyloxy)-5,11-dioxido-16-oxo-14-[(propanoyloxy)methyl]-4,6,10,12,15-pentaoxa-5,11-diphosphanonadec-1-yl undecanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, COPPER (II) ION, ... | | Authors: | Berndtsson, J, Rathore, S, Ott, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Embo Rep., 21, 2020
|
|
7SSX
 
 | | Structure of human Kv1.3 | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSZ
 
 | | Structure of human Kv1.3 with A0194009G09 nanobodies | | Descriptor: | Nanobody A0194009G09, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3,Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
