3Q6G
 
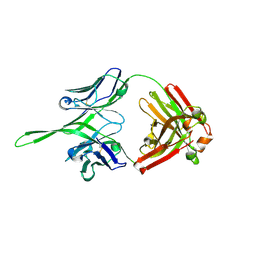 | | Crystal structure of Fab of rhesus mAb 2.5B specific for quaternary neutralizing epitope of HIV-1 gp120 | | Descriptor: | Heavy chain of Fab of rhesus mAb 2.5B, Light chain of Fab of rhesus mAb 2.5B | | Authors: | Spurrier, B, Sampson, J, Totrov, M, Li, H, O'Neal, T, William, C, Robinson, J, Gorny, M.K, Zolla-Pazner, S, Kong, X.P. | | Deposit date: | 2010-12-31 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Analysis of Human and Macaque mAbs 2909 and 2.5B: Implications for the Configuration of the Quaternary Neutralizing Epitope of HIV-1 gp120.
Structure, 19, 2011
|
|
7K3M
 
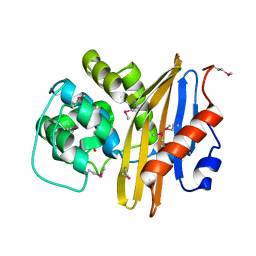 | | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography | | Descriptor: | Beta-lactamase | | Authors: | Kim, Y, Sherrell, D.A, Johnson, J, Lavens, A, Maltseva, N, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-11 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography
To Be Published
|
|
3F64
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-O)paranitrophenyl ligand | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, F17a-G | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
1ZEI
 
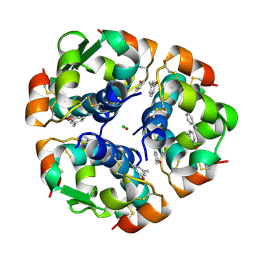 | | CROSS-LINKED B28 ASP INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-07-14 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
6KVQ
 
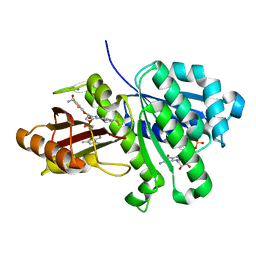 | | S. aureus FtsZ in complex with BOFP (compound 3) | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ferrer-Gonzalez, E, Fujita, J, Yoshizawa, T, Nelson, J.M, Pilch, A.J, Hillman, E, Ozawa, M, Kuroda, N, Parhi, A.K, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2019-09-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens.
Sci Rep, 9, 2019
|
|
7CRN
 
 | |
3F2E
 
 | | Crystal structure of Yellowstone SIRV coat protein C-terminus | | Descriptor: | CITRIC ACID, SIRV coat protein | | Authors: | Taurog, R.E, Szymczyna, B.R, Williamson, J.R, Johnson, J.E. | | Deposit date: | 2008-10-29 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | Synergy of NMR, computation, and X-ray crystallography for structural biology.
Structure, 17, 2009
|
|
1ZEG
 
 | | STRUCTURE OF B28 ASP INSULIN IN COMPLEX WITH PHENOL | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
4NPE
 
 | | High-resolution structure of C domain of staphylococcal protein A at room temperature | | Descriptor: | Immunoglobulin G-binding protein A, THIOCYANATE ION, ZINC ION | | Authors: | Deis, L.N, Pemble IV, C.W, Oas, T.G, Richardson, J.S, Richardson, D.C. | | Deposit date: | 2013-11-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Multiscale conformational heterogeneity in staphylococcal protein a: possible determinant of functional plasticity.
Structure, 22, 2014
|
|
6KVP
 
 | | S. aureus FtsZ in complex with 3-(1-(5-bromo-4-(4-(trifluoromethyl)phenyl)oxazol-2-yl)ethoxy)-2,6-difluorobenzamide (compound 2) | | Descriptor: | 3-[(1R)-1-[5-bromanyl-4-[4-(trifluoromethyl)phenyl]-1,3-oxazol-2-yl]ethoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Ferrer-Gonzalez, E, Fujita, J, Yoshizawa, T, Nelson, J.M, Pilch, A.J, Hillman, E, Ozawa, M, Kuroda, N, Parhi, A.K, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2019-09-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens.
Sci Rep, 9, 2019
|
|
3F6J
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-3)Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, F17a-G | | Authors: | Buts, L, de Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli
Biology, 2, 2013
|
|
3FFO
 
 | | F17b-G lectin domain with bound GlcNAc(beta1-2)man | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose, Adhesin, ... | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-12-04 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
1ZEH
 
 | | STRUCTURE OF INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Whittingham, J.L, Edwards, E.J, Antson, A.A, Clarkson, J.M, Dodson, G.G. | | Deposit date: | 1998-05-01 | | Release date: | 1998-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions of phenol and m-cresol in the insulin hexamer, and their effect on the association properties of B28 pro --> Asp insulin analogues.
Biochemistry, 37, 1998
|
|
6B2U
 
 | | Crystal structure of Xanthomonas campestris OleA H285N with Cerulenin | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, 3-oxoacyl-[ACP] synthase III, ... | | Authors: | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | Deposit date: | 2017-09-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The role of OleA His285 in substrate coordination of long-chain acyl-CoA
To be published
|
|
1M57
 
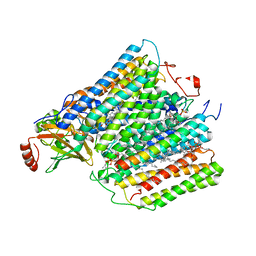 | | Structure of cytochrome c oxidase from Rhodobacter sphaeroides (EQ(I-286) mutant)) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Svensson-Ek, M, Abramson, J, Larsson, G, Tornroth, S, Brezezinski, P, Iwata, S. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides.
J.Mol.Biol., 321, 2002
|
|
5TC0
 
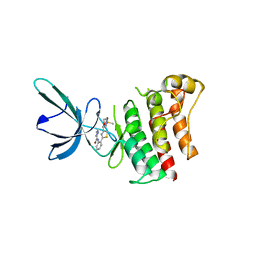 | |
5TD2
 
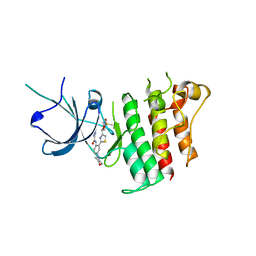 | |
5TIH
 
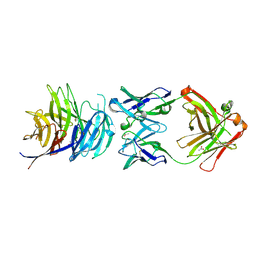 | | Structural basis for inhibition of erythrocyte invasion by antibodies to Plasmodium falciparum protein CyRPA | | Descriptor: | ACETATE ION, CyRPA antibody Fab Heavy Chain, CyRPA antibody Fab Light Chain, ... | | Authors: | Chen, L, Xu, Y, Wang, W, Thompson, J.K, Goddard-Borger, E, Lawrence, M.C, Cowman, A.F. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis for inhibition of erythrocyte invasion by antibodies toPlasmodium falciparumprotein CyRPA.
Elife, 6, 2017
|
|
5TIK
 
 | | Structural basis for inhibition of erythrocyte invasion by antibodies to Plasmodium falciparum protein CyRPA | | Descriptor: | Cysteine-rich protective antigen | | Authors: | Chen, L, Xu, Y, Wang, W, Thompson, J.K, Goddard-Borger, E, Lawrence, M.C, Cowman, A.F. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural basis for inhibition of erythrocyte invasion by antibodies toPlasmodium falciparumprotein CyRPA.
Elife, 6, 2017
|
|
5TMH
 
 | | Structure of Zika virus NS5 | | Descriptor: | GLYCEROL, Polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, B, Tan, X, Song, J. | | Deposit date: | 2016-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.278 Å) | | Cite: | The structure of Zika virus NS5 reveals a conserved domain conformation.
Nat Commun, 8, 2017
|
|
8QVH
 
 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | 4-[(trans-4-{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbamoyl]amino}cyclohexyl)oxy]benzoic acid, Bifunctional epoxide hydrolase 2 | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8QVF
 
 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | 1-(1-adamantyl)-3-(1-methylsulfonylpiperidin-4-yl)urea, Bifunctional epoxide hydrolase 2 | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8QWG
 
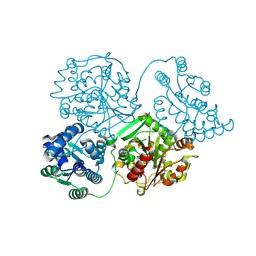 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | Bifunctional epoxide hydrolase 2, TRIETHYLENE GLYCOL | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8QVL
 
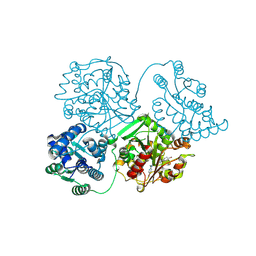 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | 2-[(5-BROMO-2-PYRIDYL)-METHYL-AMINO]ETHANOL, Bifunctional epoxide hydrolase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
8QVG
 
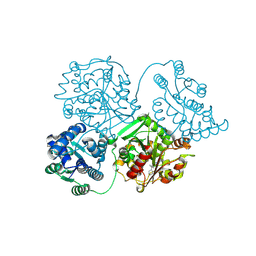 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | Bifunctional epoxide hydrolase 2, N-(3,3-DIPHENYLPROPYL)PYRROLIDINE-1-CARBOXAMIDE | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 11, 2024
|
|
