7BZU
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5YGH
 
 | | Crystal Structure of the Capsid Protein from Zika Virus | | Descriptor: | Capsid protein | | Authors: | Shang, Z, Song, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-09-23 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Crystal Structure of the Capsid Protein from Zika Virus.
J. Mol. Biol., 430, 2018
|
|
7CDZ
 
 | |
7CE0
 
 | |
5Z2Q
 
 | | Vgll1-TEAD4 core complex | | Descriptor: | PHOSPHATE ION, Transcription cofactor vestigial-like protein 1, Transcriptional enhancer factor TEF-3 | | Authors: | Pobbati, A.V, Song, H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural and functional similarity between the Vgll1-TEAD and the YAP-TEAD complexes.
Structure, 20, 2012
|
|
6LG2
 
 | | VanR bound to Vanillate | | Descriptor: | 4-HYDROXY-3-METHOXYBENZOATE, Predicted transcriptional regulators | | Authors: | He, Y, Bharath, S.R, Song, H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Developing a highly efficient hydroxytyrosol whole-cell catalyst by de-bottlenecking rate-limiting steps.
Nat Commun, 11, 2020
|
|
6KY8
 
 | | Crystal structure of ASFV dUTPase | | Descriptor: | E165R | | Authors: | Li, C, Chai, Y, Song, H, Qi, J, Sun, Y, Gao, G.F. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target.
Mbio, 10, 2019
|
|
5XSQ
 
 | | Crystal Structure of the Marburg Virus Nucleoprotein Core Domain Chaperoned by a VP35 Peptide | | Descriptor: | Nucleoprotein, Peptide from Polymerase cofactor VP35 | | Authors: | Zhu, T, Song, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-06-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Marburg Virus Nucleoprotein Core Domain Chaperoned by a VP35 Peptide Reveals a Conserved Drug Target for Filovirus
J. Virol., 91, 2017
|
|
5DMR
 
 | |
5ZMC
 
 | | Structural Basis for Reactivation of -146C>T Mutant TERT Promoter by cooperative binding of p52 and ETS1/2 | | Descriptor: | DNA (5'-D(P*CP*GP*GP*GP*GP*AP*CP*CP*CP*GP*GP*AP*AP*GP*GP*G)-3'), DNA (5'-D(P*GP*CP*CP*CP*TP*TP*CP*CP*GP*GP*GP*TP*CP*CP*CP*C)-3'), Nuclear factor NF-kappa-B p100 subunit, ... | | Authors: | Xu, X, Bharath, S.R, Song, H. | | Deposit date: | 2018-04-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for reactivating the mutant TERT promoter by cooperative binding of p52 and ETS1.
Nat Commun, 9, 2018
|
|
2LT7
 
 | | Solution NMR structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5GN0
 
 | | Structure of TAZ-TEAD complex | | Descriptor: | CITRIC ACID, PALMITIC ACID, Transcriptional enhancer factor TEF-3, ... | | Authors: | Kaan, H.Y.K, Song, H. | | Deposit date: | 2016-07-18 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of TAZ-TEAD complex reveals a distinct interaction mode from that of YAP-TEAD complex
Sci Rep, 7, 2017
|
|
5HS3
 
 | | Human thymidylate synthase complexed with dUMP and 3-amino-2-benzoyl-4-methylthieno[2,3-b]pyridin-6-ol | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 3-amino-2-benzoyl-4-methylthieno[2,3-b]pyridin-6-ol, Thymidylate synthase | | Authors: | Chen, D, Almqvist, H, Axelsson, H, Jafari, R, Mateus, A, Haraldsson, M, Larsson, A, Artursson, P, Molina, D.M, Lundback, T, Nordlund, P. | | Deposit date: | 2016-01-25 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluorouracil
Nat Commun, 7, 2016
|
|
5BUS
 
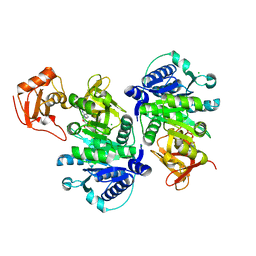 | | O-succinylbenzoate Coenzyme A Synthetase (MenE) from Bacillus Subtilis, in complex with AMP | | Descriptor: | 2-succinylbenzoate--CoA ligase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION | | Authors: | Chen, Y, Sun, Y, Song, H, Guo, Z. | | Deposit date: | 2015-06-04 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural Basis for the ATP-dependent Configuration of Adenylation Active Site in Bacillus subtilis o-Succinylbenzoyl-CoA Synthetase
J.Biol.Chem., 290, 2015
|
|
5DMQ
 
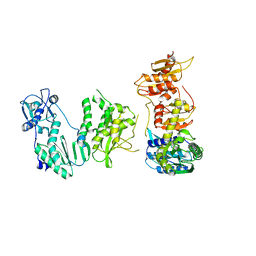 | |
1PKD
 
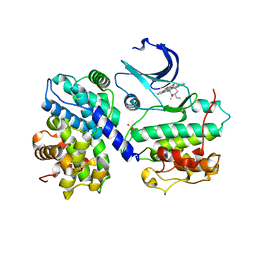 | |
5GS6
 
 | | Full-length NS1 structure of Zika virus from 2015 Brazil strain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NS1 of Zika virus from 2015 Brazil strain | | Authors: | Xu, X.Y, Song, H, Qi, J.X, Shi, Y, Gao, G.F. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Contribution of intertwined loop to membrane association revealed by Zika virus full-length NS1 structure
Embo J., 35, 2016
|
|
5BUR
 
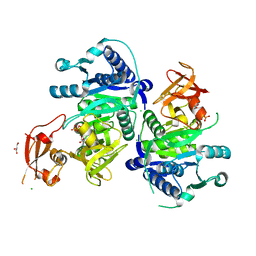 | | O-succinylbenzoate Coenzyme A Synthetase (MenE) from Bacillus Subtilis, in Complex with ATP and Magnesium Ion | | Descriptor: | 1,2-ETHANEDIOL, 2-succinylbenzoate--CoA ligase, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, Y, Sun, Y, Song, H, Guo, Z. | | Deposit date: | 2015-06-04 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Basis for the ATP-dependent Configuration of Adenylation Active Site in Bacillus subtilis o-Succinylbenzoyl-CoA Synthetase
J.Biol.Chem., 290, 2015
|
|
5JXE
 
 | | Human PD-1 ectodomain complexed with Pembrolizumab Fab | | Descriptor: | Pembrolizumab Fab heavy chain, Pembrolizumab Fab light chain, Programmed cell death protein 1 | | Authors: | Na, Z, Bharath, S.R, Song, H. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for blocking PD-1-mediated immune suppression by therapeutic antibody pembrolizumab.
Cell Res., 27, 2017
|
|
7BZO
 
 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4Z
 
 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4Y
 
 | | Cryo-EM structure of empty Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4T
 
 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7C4W
 
 | | Cryo-EM structure of A particle Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6DFA
 
 | | Kaiso (ZBTB33) E535A zinc finger DNA binding domain in complex with the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | A conformational switch in the zinc finger protein Kaiso mediates differential readout of specific and methylated DNA sequences.
Biochemistry, 2020
|
|
