7M3G
 
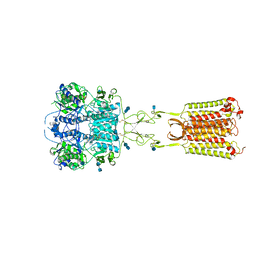 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-[4-[(3S)-3-[[(1R)-1-naphthalen-1-ylethyl]amino]pyrrolidin-1-yl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
7M3E
 
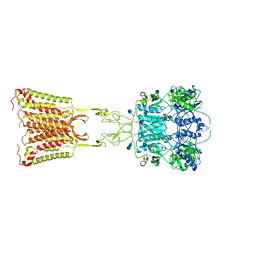 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
7M3J
 
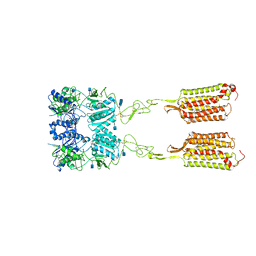 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
7M3F
 
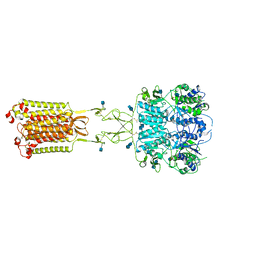 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
6XEU
 
 | | CryoEM structure of GIRK2PIP2* - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2 | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, SODIUM ION, ... | | Authors: | Mathiharan, Y.K, Glaaser, I.W, Skiniotis, G, Slesinger, P.A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into GIRK2 channel modulation by cholesterol and PIP2
Cell Rep, 36, 2021
|
|
6XEV
 
 | | CryoEM structure of GIRK2-PIP2/CHS - G protein-gated inwardly rectifying potassium channel GIRK2 with modulators cholesteryl hemisuccinate and PIP2 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, ... | | Authors: | Mathiharan, Y.K, Glaaser, I.W, Skiniotis, G, Slesinger, P.A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into GIRK2 channel modulation by cholesterol and PIP2
Cell Rep, 36, 2021
|
|
4X8P
 
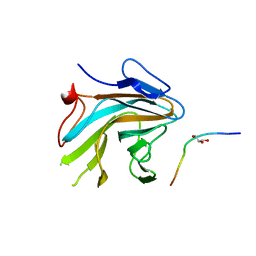 | | Crystal structure of Ash2L SPRY domain in complex with RbBP5 | | Descriptor: | GLYCEROL, Retinoblastoma-binding protein 5, Set1/Ash2 histone methyltransferase complex subunit ASH2,Set1/Ash2 histone methyltransferase complex subunit ASH2 | | Authors: | Zhang, P, Chaturvedi, C.P, Brunzelle, J.S, Skiniotis, G, Brand, M, Shilatifard, A, Couture, J.-F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation.
Genes Dev., 29, 2015
|
|
4X8N
 
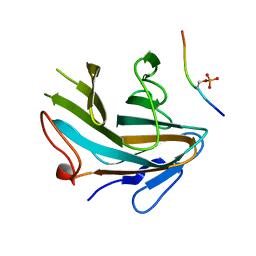 | | Crystal structure of Ash2L SPRY domain in complex with phosphorylated RbBP5 | | Descriptor: | Retinoblastoma-binding protein 5, Set1/Ash2 histone methyltransferase complex subunit ASH2 | | Authors: | Zhang, P, Chaturvedi, C.P, Brunzelle, J.S, Skiniotis, G, Brand, M, Shilatifard, A, Couture, J.-F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation.
Genes Dev., 29, 2015
|
|
2V5S
 
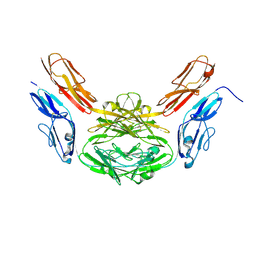 | | Structural basis for Dscam isoform specificity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DSCAM | | Authors: | Meijers, R, Puettmann-Holgado, R, Skiniotis, G, Liu, J.-H, Walz, T, Schmucker, D, Wang, J.-H. | | Deposit date: | 2007-07-09 | | Release date: | 2007-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Dscam Isoform Specificity
Nature, 449, 2007
|
|
7UL5
 
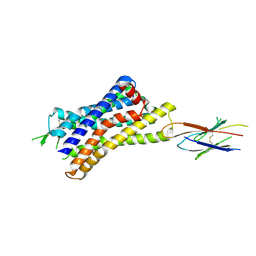 | |
7UL3
 
 | | CryoEM Structure of Inactive H2R Bound to Famotidine, Nb6M, and NabFab | | Descriptor: | Histamine H2 receptor, NabFab HC, NabFab LC, ... | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UL2
 
 | | CryoEM Structure of Inactive NTSR1 Bound to SR48692 and Nb6 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Nanobody 6, Neurotensin receptor 1, ... | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UL4
 
 | | CryoEM Structure of Inactive MOR Bound to Alvimopan and Mb6 | | Descriptor: | Megabody 6, Mu-type opioid receptor, N-[(2S)-2-{[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]methyl}-3-phenylpropanoyl]glycine | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2V5R
 
 | | Structural basis for Dscam isoform specificity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DSCAM, GLYCEROL | | Authors: | Meijers, R, Puettmann-Holgado, R, Skiniotis, G, Liu, J.-H, Walz, T, Schmucker, D, Wang, J.-H. | | Deposit date: | 2007-07-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Dscam Isoform Specificity
Nature, 449, 2007
|
|
5UZ7
 
 | | Volta phase plate cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | Descriptor: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liang, Y.L, Khoshouei, M, Radjainia, M, Zhang, Y, Glukhova, A, Tarrasch, J, Thal, D.M, Furness, S.G.B, Christopoulos, G, Coudrat, T, Danev, R, Baumeister, W, Miller, L.J, Christopoulos, A, Kobilka, B.K, Wootten, D, Skiniotis, G, Sexton, P.M. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Phase-plate cryo-EM structure of a class B GPCR-G-protein complex.
Nature, 546, 2017
|
|
2V5M
 
 | | Structural basis for Dscam isoform specificity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DSCAM, GLYCEROL | | Authors: | Meijers, R, Puettmann-Holgado, R, Skiniotis, G, Liu, J.-H, Walz, T, Schmucker, D, Wang, J.-H. | | Deposit date: | 2007-07-06 | | Release date: | 2007-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis of Dscam Isoform Specificity
Nature, 449, 2007
|
|
5VAI
 
 | | Cryo-EM structure of the activated Glucagon-like peptide-1 receptor in complex with G protein | | Descriptor: | Glucagon-like peptide 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, Y, Sun, B, Feng, D, Hu, H, Chu, M, Qu, Q, Tarrasch, J.T, Li, S, Kobilka, T.S, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2017-03-27 | | Release date: | 2017-05-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein.
Nature, 546, 2017
|
|
5UQ7
 
 | | 70S ribosome complex with dnaX mRNA stemloop and E-site tRNA ("in" conformation) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | Deposit date: | 2017-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
5UQ8
 
 | | 70S ribosome complex with dnaX mRNA stem-loop and E-site tRNA ("out" conformation) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | Deposit date: | 2017-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2018-03-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
6N4X
 
 | | Metabotropic Glutamate Receptor 5 Apo Form Ligand Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N51
 
 | | Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N52
 
 | | Metabotropic Glutamate Receptor 5 Apo Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N4B
 
 | | Cannabinoid Receptor 1-G Protein Complex | | Descriptor: | CHOLESTEROL, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Krishna Kumar, K, Shalev-Benami, M, Hu, H, Weis, W.I, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2018-11-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a Signaling Cannabinoid Receptor 1-G Protein Complex.
Cell, 176, 2019
|
|
6OIJ
 
 | | Muscarinic acetylcholine receptor 1-G11 protein complex | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Antibody fragment, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maeda, S, Qianhui, Q, Skiniotis, G, Kobilka, B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-08 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes.
Science, 364, 2019
|
|
6OYA
 
 | | Structure of the Rhodopsin-Transducin-Nanobody Complex | | Descriptor: | Camelid antibody VHH fragment, Gt-alpha/Gi1-alpha chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gao, Y, Hu, H, Ramachandran, S, Erickson, J.W, Cerione, R.A, Skiniotis, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation.
Mol.Cell, 75, 2019
|
|
