6G1O
 
 | |
2YCB
 
 | | Structure of the archaeal beta-CASP protein with N-terminal KH domains from Methanothermobacter thermautotrophicus | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Silva, A.P.G, Chechik, M, Byrne, R.T, Waterman, D.G, Ng, C.L, Dodson, E.J, Koonin, E.V, Antson, A.A, Smits, C. | | Deposit date: | 2011-03-13 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and Activity of a Novel Archaeal Beta-Casp Protein with N-Terminal Kh Domains.
Structure, 19, 2011
|
|
2L3I
 
 | | Oxki4a, spider derived antimicrobial peptide | | Descriptor: | AOXKI4A, antimicrobial peptide in spider venom | | Authors: | Vassilevski, A.A, Dubovskii, P.V, Samsonova, O.V, Egorova, N.S, Kozlov, S.A, Feofanov, A.V, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2010-09-14 | | Release date: | 2011-09-14 | | Last modified: | 2019-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel lynx spider toxin shares common molecular architecture with defense peptides from frog skin.
Febs J., 278, 2011
|
|
1SME
 
 | | PLASMEPSIN II, A HEMOGLOBIN-DEGRADING ENZYME FROM PLASMODIUM FALCIPARUM, IN COMPLEX WITH PEPSTATIN A | | Descriptor: | PLASMEPSIN II, Pepstatin | | Authors: | Silva, A.M, Lee, A.Y, Gulnik, S.V, Goldberg, D.E, Erickson, J.W. | | Deposit date: | 1996-06-11 | | Release date: | 1997-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
2N5N
 
 | | Structure of an N-terminal domain of CHD4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Silva, A.P.G, Mackay, J.P. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminal Region of Chromodomain Helicase DNA-binding Protein 4 (CHD4) Is Essential for Activity and Contains a High Mobility Group (HMG) Box-like-domain That Can Bind Poly(ADP-ribose).
J.Biol.Chem., 291, 2016
|
|
4FE6
 
 | | Crystal Structure of HIV-1 Protease in Complex with an enamino-oxindole inhibitor | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[({(3Z)-3-[1-(methylamino)ethylidene]-2-oxo-2,3-dihydro-1H-indol-5-yl}sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, HIV protease | | Authors: | Silva, A.M, Eissenstat, M, Guerassina, T, Gulnik, S, Afonina, E, Yu, B, Erickson, J, Ludke, D, Yokoe, H. | | Deposit date: | 2012-05-29 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enamino-oxindole HIV protease inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1DIF
 
 | | HIV-1 PROTEASE IN COMPLEX WITH A DIFLUOROKETONE CONTAINING INHIBITOR A79285 | | Descriptor: | BETA-MERCAPTOETHANOL, HIV-1 PROTEASE, N-{1-BENZYL-2,2-DIFLUORO-3,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Silva, A.M, Cachau, R.E, Sham, H.L, Erickson, J.W. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition and catalytic mechanism of HIV-1 aspartic protease.
J.Mol.Biol., 255, 1996
|
|
7R79
 
 | | Histoplasma capsulatum H88 Calcium Binding Protein 1 (Cbp1) | | Descriptor: | Calcium-binding protein | | Authors: | Herrera, N, Azimova, D, Sil, A, Rosenberg, O.S. | | Deposit date: | 2021-06-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cbp1, a fungal virulence factor under positive selection, forms an effector complex that drives macrophage lysis.
Plos Pathog., 18, 2022
|
|
7R6U
 
 | | Paracoccidioides americana Pb03 Calcium Binding Protein 1 (Cbp1) | | Descriptor: | CBP domain-containing protein | | Authors: | Herrera, N, Azimova, D, Sil, A, Rosenberg, O.S. | | Deposit date: | 2021-06-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cbp1, a fungal virulence factor under positive selection, forms an effector complex that drives macrophage lysis.
Plos Pathog., 18, 2022
|
|
6CME
 
 | | Structure of wild-type ISL2-LID in complex with LHX4-LIM1+2 | | Descriptor: | LIM/homeobox protein Lhx4,Insulin gene enhancer protein ISL-2, ZINC ION | | Authors: | Stokes, P.H, Silva, A, Guss, J.M, Matthews, J.M. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins, 87, 2019
|
|
6W6V
 
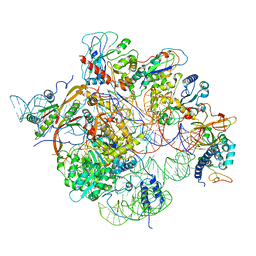 | | Structure of yeast RNase MRP holoenzyme | | Descriptor: | RNA component of RNase MRP NME1, RNases MRP/P 32.9 kDa subunit, Ribonuclease MRP protein subunit RMP1, ... | | Authors: | Perederina, A, Li, D, Lee, H, Bator, C, Berezin, I, Hafenstein, S.L, Krasilnikov, A.S. | | Deposit date: | 2020-03-17 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of catalytic ribonucleoprotein complex RNase MRP.
Nat Commun, 11, 2020
|
|
8AA3
 
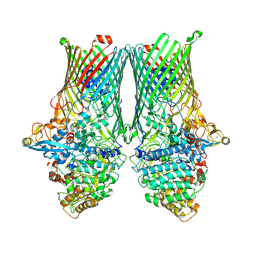 | | Core SusCD transporter units from the inactive levan utilisome in the presence of levan fructo-oligosaccharides DP 15-25 | | Descriptor: | MAGNESIUM ION, SusC homolog, SusD homolog, ... | | Authors: | White, J.B.R, Silale, A, Ranson, N.A, van den Berg, B. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes.
Nature, 618, 2023
|
|
8A9Y
 
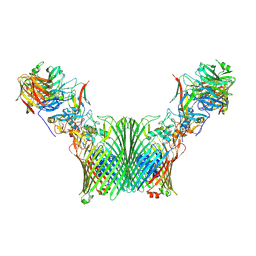 | | Substrate-free levan utilisation machinery (utilisome) | | Descriptor: | DUF4960 domain-containing protein, Glycoside hydrolase family 32, MAGNESIUM ION, ... | | Authors: | White, J.B.R, Silale, A, Ranson, N.A, van den Berg, B. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes.
Nature, 618, 2023
|
|
8AA1
 
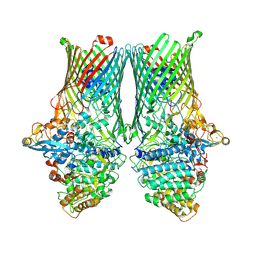 | | Core SusCD transporter units from the levan utilisome with levan fructo-oligosaccharides DP 8-12 | | Descriptor: | MAGNESIUM ION, SusC homolog, SusD homolog, ... | | Authors: | White, J.B.R, Silale, A, Ranson, N.A, van den Berg, B. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes.
Nature, 618, 2023
|
|
8AA0
 
 | | Levan utilisation machinery (utilisome) with levan fructo-oligosaccharides DP 8-12 | | Descriptor: | DUF4960 domain-containing protein, Glycoside hydrolase family 32, MAGNESIUM ION, ... | | Authors: | White, J.B.R, Silale, A, Ranson, N.A, van den Berg, B. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes.
Nature, 618, 2023
|
|
8AA2
 
 | | Inactive levan utilisation machinery (utilisome) in the presence of levan fructo-oligosaccharides DP 15-25 | | Descriptor: | DUF4960 domain-containing protein, Glycoside hydrolase family 32, MAGNESIUM ION, ... | | Authors: | White, J.B.R, Silale, A, Ranson, N.A, van den Berg, B. | | Deposit date: | 2022-06-29 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes.
Nature, 618, 2023
|
|
8AA4
 
 | |
4RCT
 
 | | Crystal structure of R-protein of NgoAVII restriction endonuclease | | Descriptor: | Restriction endonuclease R.NgoVII | | Authors: | Tamulaitiene, G, Silanskas, A, Grazulis, S, Zaremba, M, Siksnys, V. | | Deposit date: | 2014-09-17 | | Release date: | 2014-12-24 | | Last modified: | 2014-12-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the R-protein of the multisubunit ATP-dependent restriction endonuclease NgoAVII.
Nucleic Acids Res., 42, 2014
|
|
4RD5
 
 | | Crystal structure of R.NgoAVII restriction endonuclease B3 domain with cognate DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*TP*AP*AP*GP*CP*GP*GP*CP*AP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*TP*TP*GP*CP*CP*GP*CP*TP*TP*AP*GP*G)-3'), Restriction endonuclease R.NgoVII | | Authors: | Tamulaitiene, G, Silanskas, A, Grazulis, S, Zaremba, M, Siksnys, V. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the R-protein of the multisubunit ATP-dependent restriction endonuclease NgoAVII.
Nucleic Acids Res., 42, 2014
|
|
4RDM
 
 | | Crystal structure of R.NgoAVII restriction endonuclease B3 domain with cognate DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*AP*GP*CP*GP*GP*CP*AP*AP*TP*CP*C)-3)', DNA (5'-D(*GP*GP*GP*AP*TP*TP*GP*CP*CP*GP*CP*TP*TP*AP*G)-3)', Restriction endonuclease R.NgoVII, ... | | Authors: | Tamulaitiene, G, Silanskas, A, Grazulis, S, Zaremba, M, Siksnys, V. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the R-protein of the multisubunit ATP-dependent restriction endonuclease NgoAVII.
Nucleic Acids Res., 42, 2014
|
|
4WGF
 
 | | YcaC from Pseudomonas aeruginosa with hexane-2,5-diol and covalent acrylamide | | Descriptor: | (2R,5R)-hexane-2,5-diol, CHLORIDE ION, PROPIONAMIDE, ... | | Authors: | Groftehauge, M.K, Truan, D, Vasil, A, Denny, P.W, Vasil, M.L, Pohl, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34020686 Å) | | Cite: | Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide Adduct.
Int J Mol Sci, 16, 2015
|
|
4WH0
 
 | | YcaC from Pseudomonas aeruginosa with S-mercaptocysteine active site cysteine | | Descriptor: | CHLORIDE ION, Putative hydrolase | | Authors: | Groftehauge, M.K, Truan, D, Vasil, A, Denny, P.W, Vasil, M.L, Pohl, E. | | Deposit date: | 2014-09-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.563 Å) | | Cite: | Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide Adduct.
Int J Mol Sci, 16, 2015
|
|
4UIF
 
 | | Cryo-EM structure of Dengue virus serotype 2 in complex with antigen-binding fragments of human antibody 2D22 | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - HEAVY CHAIN, ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - LIGHT CHAIN, DENGUE VIRUS SEROTYPE 2 STRAIN PVP94 07 - ENVELOPE PROTEIN, ... | | Authors: | Fibriansah, G, Ibarra, K.D, Ng, T.-S, Smith, S.A, Tan, J.L, Lim, X.-N, Ooi, J.S.G, Kostyuchenko, V.A, Wang, J, de Silva, A.M, Harris, E, Crowe Junior, J.E, Lok, S.-M. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers.
Science, 349, 2015
|
|
4UIH
 
 | | Cryo-EM structure of Dengue virus serotype 2 strain New Guinea-C complexed with human antibody 2D22 Fab at 37 degree C. The Fab molecules were added to the virus before 37 degree C incubation. | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 -HEAVY CHAIN, ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 -LIGHT CHAIN, DENGUE VIRUS SEROTYPE 2 STRAIN NEW GUINEA-C E PROTEIN ECTODOMAIN | | Authors: | Fibriansah, G, Ibarra, K.D, Ng, T.-S, Smith, S.A, Tan, J.L, Lim, X.N, Ooi, J.S.G, Kostyuchenko, V.A, Wang, J, de Silva, A.M, Harris, E, Crowe, J.E, Lok, S.-M. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers.
Science, 349, 2015
|
|
6V6H
 
 | | Crystal structure of histidine ammonia-lyase from Trypanosoma cruzi | | Descriptor: | Histidine ammonia-lyase | | Authors: | Miranda, R.R, Silva, M, Barison, M.J, Silber, A.M, Iulek, J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of histidine ammonia-lyase from Trypanosoma cruzi.
Biochimie, 175, 2020
|
|
