3O86
 
 | | Crystal structure of AmpC beta-lactamase in complex with a sulfonamide boronic acid inhibitor | | Descriptor: | Beta-lactamase, PHOSPHATE ION, {[(benzylsulfonyl)amino]methyl}boronic acid | | Authors: | Eidam, O, Romagnoli, C, Karpiak, J, Shoichet, B.K. | | Deposit date: | 2010-08-02 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, Synthesis, Crystal Structures, and Antimicrobial Activity of Sulfonamide Boronic Acids as beta-Lactamase Inhibitors
J.Med.Chem., 53, 2010
|
|
3O88
 
 | | Crystal structure of AmpC beta-lactamase in complex with a sulfonamide boronic acid inhibitor | | Descriptor: | 3-[(2R)-2-[(benzylsulfonyl)amino]-2-(dihydroxyboranyl)ethyl]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Romagnoli, C, Karpiak, J, Shoichet, B.K. | | Deposit date: | 2010-08-02 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design, Synthesis, Crystal Structures, and Antimicrobial Activity of Sulfonamide Boronic Acids as beta-Lactamase Inhibitors
J.Med.Chem., 53, 2010
|
|
5WIV
 
 | | Structure of the sodium-bound human D4 Dopamine receptor in complex with Nemonapride | | Descriptor: | D(4) dopamine receptor, soluble cytochrome b562 chimera, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wacker, D, Wang, S, Levit, A, Che, T, Betz, R.M, McCorvy, J.D, Venkatakrishnan, A.J, Huang, X.-P, Dror, R.O, Shoichet, B.K, Roth, B.L. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | D4 dopamine receptor high-resolution structures enable the discovery of selective agonists.
Science, 358, 2017
|
|
7W6P
 
 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a G protein biased agonist | | Descriptor: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
7W7E
 
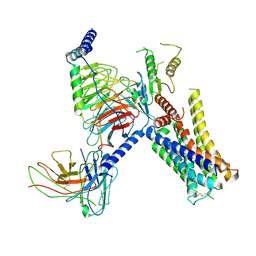 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a biased agonist | | Descriptor: | 5-(3-bicyclo[4.2.0]octa-1,3,5-trienyl)-1,2,3,6-tetrahydropyridine, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
5WIU
 
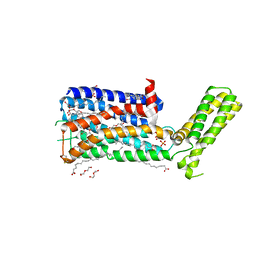 | | Structure of the human D4 Dopamine receptor in complex with Nemonapride | | Descriptor: | D(4) dopamine receptor, soluble cytochrome b562 chimera, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wacker, D, Wang, S, Levit, A, Che, T, Betz, R.M, McCorvy, J.D, Venkatakrishnan, A.J, Huang, X.-P, Dror, R.O, Shoichet, B.K, Roth, B.L. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | D4 dopamine receptor high-resolution structures enable the discovery of selective agonists.
Science, 358, 2017
|
|
5JWV
 
 | | T4 Lysozyme L99A/M102Q with Ethylbenzene Bound | | Descriptor: | Endolysin, PHENYLETHANE | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWT
 
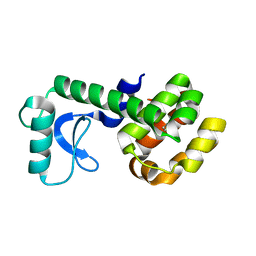 | | T4 Lysozyme L99A/M102Q with Benzene Bound | | Descriptor: | BENZENE, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWU
 
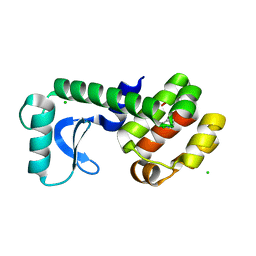 | | T4 Lysozyme L99A/M102Q with 1,2-Dihydro-1,2-azaborine Bound | | Descriptor: | 1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
9C81
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | (2R)-2-phenoxy-3-{[(1S,2S,4S)-spiro[bicyclo[2.2.1]heptane-7,1'-cyclopropane]-2-carbonyl]amino}propanoic acid, AmpC Beta-lactamase | | Authors: | Liu, F, Shoichet, B.K, Bassim, V. | | Deposit date: | 2024-06-11 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
9C6P
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | 3-chloro-N-(5-chloro-2-methyl-1,3-benzothiazol-6-yl)-2-hydroxybenzene-1-sulfonamide, AmpC Beta-lactamase | | Authors: | Liu, F, Shoichet, B.K. | | Deposit date: | 2024-06-08 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.663 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
9C8J
 
 | |
9C83
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | AmpC Beta-lactamase, N-[(3M)-3-(5-chloro-1,2,3-thiadiazol-4-yl)phenyl]-5-methyl-3-oxo-2,3-dihydro-1,2-oxazole-4-sulfonamide | | Authors: | Liu, F, Shoichet, B.K. | | Deposit date: | 2024-06-11 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
6U09
 
 | |
9C84
 
 | |
6U06
 
 | |
2HDQ
 
 | |
2I72
 
 | | AmpC beta-lactamase in complex with 5-diformylaminomethyl-benzo[b]thiophen-2-boronic acid | | Descriptor: | Beta-lactamase, {5-[(DIFORMYLAMINO)METHYL]-1-BENZOTHIEN-2-YL}BORONIC ACID | | Authors: | Venturelli, A, Cancian, L, Tondi, D, Morandi, F, Cannazza, G, Segatore, B, Prati, F, Amicosante, G, Shoichet, B.K, Costi, M.P. | | Deposit date: | 2006-08-30 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimizing Cell Permeation of an Antibiotic Resistance Inhibitor for Improved Efficacy
J.Med.Chem., 50, 2007
|
|
2HDR
 
 | |
2HDS
 
 | |
2HDU
 
 | |
5A7W
 
 | | Crystal structure of human JMJD2A in complex with compound 35 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[(4-hydroxyphenyl)carbonylamino]-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5A7S
 
 | | Crystal structure of human JMJD2A in complex with compound 44 | | Descriptor: | 1,2-ETHANEDIOL, 2-(5-acetamido-2-oxidanyl-phenyl)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
8DIC
 
 | |
8DIF
 
 | |
