6ON2
 
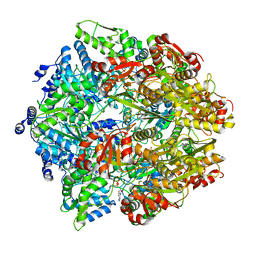 | | Lon Protease from Yersinia pestis with Y2853 substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent protease La, ... | | Authors: | Shin, M, Asmita, A, Puchades, C, Adjei, E, Wiseman, R.L, Karzai, A.W, Lander, G.C. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for distinct operational modes and protease activation in AAA+ protease Lon.
Sci Adv, 6, 2020
|
|
6V11
 
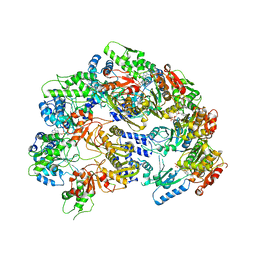 | | Lon Protease from Yersinia pestis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Shin, M, Puchades, C, Asmita, A, Puri, N, Adjei, E, Wiseman, R.L, Karzai, A.W, Lander, G.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for distinct operational modes and protease activation in AAA+ protease Lon.
Sci Adv, 6, 2020
|
|
7KSM
 
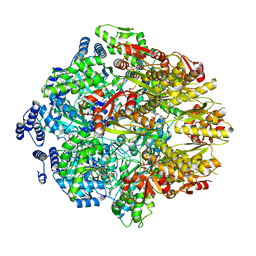 | | Human mitochondrial LONP1 with endogenous substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7KRZ
 
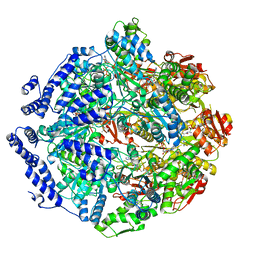 | | Human mitochondrial LONP1 in complex with Bortezomib | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Endogenous co-purified substrate, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-20 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7KSL
 
 | | Substrate-free human mitochondrial LONP1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
209D
 
 | | Structural, physical and biological characteristics of RNA:DNA binding agent N8-actinomycin D | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3'), N8-ACTINOMYCIN D | | Authors: | Shinomiya, M, Chu, W, Carlson, R.G, Weaver, R.F, Takusagawa, F. | | Deposit date: | 1995-05-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural, Physical, and Biological Characteristics of RNA.DNA Binding Agent N8-Actinomycin D.
Biochemistry, 34, 1995
|
|
4WUA
 
 | | Crystal structure of human SRPK1 complexed to an inhibitor SRPIN340 | | Descriptor: | CITRIC ACID, N-[2-(1-piperidinyl)-5-(trifluoromethyl)phenyl]-4-pyridinecarboxamide, SRSF protein kinase 1, ... | | Authors: | Hoshina, M, Ikura, T, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Dual Inhibitor of SRPK1 and CK2 That Attenuates Pathological Angiogenesis of Macular Degeneration in Mice
Mol.Pharmacol., 88, 2015
|
|
3KPX
 
 | | Crystal Structure Analysis of photoprotein clytin | | Descriptor: | Apophotoprotein clytin-3, C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION | | Authors: | Titushin, M.S, Li, Y, Stepanyuk, G.A, Wang, B.-C, Lee, J, Vysotski, E.S, Liu, Z.-J. | | Deposit date: | 2009-11-17 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | NMR derived topology of a GFP-photoprotein energy transfer complex
J.Biol.Chem., 285, 2010
|
|
7DVV
 
 | | Heme sensor protein PefR from Streptococcus agalactiae bound to operator DNA (28-mer) | | Descriptor: | DNA (28-MER), HTH marR-type domain-containing protein | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
7DVU
 
 | | Crystal structure of heme sensor protein PefR in complex with heme and cyanide | | Descriptor: | CYANIDE ION, HTH marR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
7DVT
 
 | | Crystal structure of heme sensor protein PefR in complex with heme and carbon monoxide | | Descriptor: | CARBON MONOXIDE, HTH marR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
7DVR
 
 | | Crystal structure of heme sensor protein PefR from Streptococcus agalactiae in complex with heme | | Descriptor: | COBALT (II) ION, HTH marR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
2D55
 
 | | Structural, physical and biological characteristics of RNA.DNA binding agent N8-actinomycin D | | Descriptor: | ACTINOMYCIN D, DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3') | | Authors: | Shinomiya, M, Chu, W, Carlson, R.G, Weaver, R.F, Takusagawa, F. | | Deposit date: | 1995-05-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 2:1 Complex between D(Gaagcttc) and the Anticancer Drug Actinomycin D.
J.Mol.Biol., 225, 1992
|
|
1G4F
 
 | | NMR STRUCTURE OF THE FIFTH DOMAIN OF HUMAN BETA2-GLYCOPROTEIN I | | Descriptor: | BETA2-GLYCOPROTEIN I | | Authors: | Hoshino, M, Hagihara, Y, Nishii, I, Yamazaki, T, Kato, H, Goto, Y. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Identification of the phospholipid-binding site of human beta(2)-glycoprotein I domain V by heteronuclear magnetic resonance.
J.Mol.Biol., 304, 2000
|
|
1G4G
 
 | | NMR STRUCTURE OF THE FIFTH DOMAIN OF HUMAN BETA2-GLYCOPROTEIN I | | Descriptor: | BETA2-GLYCOPROTEIN I | | Authors: | Hoshino, M, Hagihara, Y, Nishii, I, Yamazaki, T, Kato, H, Goto, Y. | | Deposit date: | 2000-10-27 | | Release date: | 2000-11-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Identification of the phospholipid-binding site of human beta(2)-glycoprotein I domain V by heteronuclear magnetic resonance.
J.Mol.Biol., 304, 2000
|
|
4L6U
 
 | | Crystal structure of AF1868: Cmr1 subunit of the Cmr RNA silencing complex | | Descriptor: | Putative uncharacterized protein | | Authors: | Sun, J, Jeon, J.H, Shin, M, Shin, H.C, Oh, B.H, Kim, J.S. | | Deposit date: | 2013-06-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and CRISPR RNA-binding site of the Cmr1 subunit of the Cmr interference complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6VN0
 
 | | BG505 SOSIP.v4.1 in complex with rhesus macaque Fab RM20F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Cottrell, C.A, Shin, M, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
3B1F
 
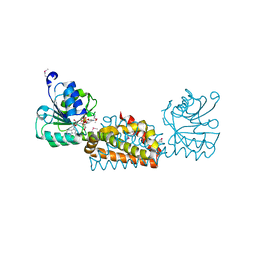 | | Crystal structure of prephenate dehydrogenase from Streptococcus mutans | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative prephenate dehydrogenase | | Authors: | Ku, H.K, Do, N.H, Song, J.S, Choi, S, Shin, M.H, Kim, K.J, Lee, S.J. | | Deposit date: | 2011-07-02 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of prephenate dehydrogenase from Streptococcus mutans.
Int.J.Biol.Macromol., 49, 2011
|
|
6AZ0
 
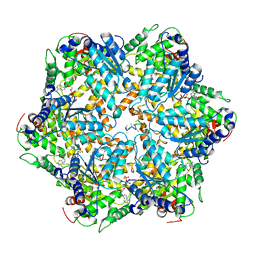 | | Mitochondrial ATPase Protease YME1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Puchades, C, Rampello, A.J, Shin, M, Giuliano, C, Wiseman, R.L, Glynn, S.E, Lander, G.C. | | Deposit date: | 2017-09-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing.
Science, 358, 2017
|
|
4Q2C
 
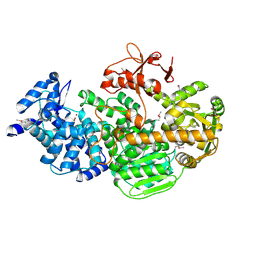 | | Crystal structure of CRISPR-associated protein | | Descriptor: | CRISPR-associated helicase Cas3, NICKEL (II) ION | | Authors: | Gong, B, Shin, M, Sun, J, van der Oost, J, Kim, J.-S. | | Deposit date: | 2014-04-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q2D
 
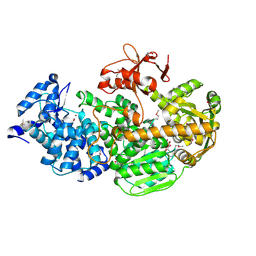 | | Crystal Structure of CRISPR-Associated protein in complex with 2'-Deoxyadenosine 5'-Triphosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CRISPR-associated helicase Cas3, MAGNESIUM ION, ... | | Authors: | Gong, B, Shin, M, Sun, J, van der Oost, J, Kim, J.-S. | | Deposit date: | 2014-04-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.771 Å) | | Cite: | Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N06
 
 | |
4GKF
 
 | | Crystal structure and characterization of Cmr5 protein from Pyrococcus furiosus | | Descriptor: | CRISPR system Cmr subunit Cmr5 | | Authors: | Park, J, Sun, J, Park, S, Hwang, H, Park, M, Shin, M.S. | | Deposit date: | 2012-08-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Cmr5 from Pyrococcus furiosus and its functional implications
Febs Lett., 587, 2013
|
|
6LNG
 
 | | Rapid crystallization of streptavidin using charged peptides | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Minamihata, K, Tsukamoto, K, Adachi, M, Shimizu, R, Mishina, M, Kuroki, R, Nagamune, T. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8000015 Å) | | Cite: | Genetically fused charged peptides induce rapid crystallization of proteins.
Chem.Commun.(Camb.), 56, 2020
|
|
6M7G
 
 | | Crystal structure of ArsN, N-acetyltransferase with substrate phosphinothricin from Pseudomonas putida KT2440 | | Descriptor: | PHOSPHINOTHRICIN, Phosphinothricin N-acetyltransferase | | Authors: | Venkadesh, S, Dheeman, D.S, Yoshinaga, M, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.657 Å) | | Cite: | Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic.
Commun Biol, 2, 2019
|
|
