4N81
 
 | |
4YGQ
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | Hydrolase, TERTIARY-BUTYL ALCOHOL | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YGR
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YGS
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | CITRIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
1YGZ
 
 | | Crystal Structure of Inorganic Pyrophosphatase from Helicobacter pylori | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Wu, C.A, Lokanath, N.K, Kim, D.Y, Park, H.J, Hwang, H.Y, Kim, S.T, Suh, S.W, Kim, K.K. | | Deposit date: | 2005-01-06 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of inorganic pyrophosphatase from Helicobacter pylori.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2MXD
 
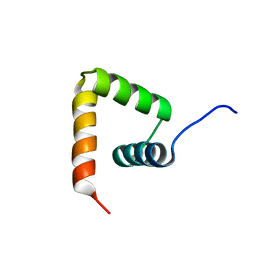 | | Solution structure of VPg of porcine sapovirus | | Descriptor: | Viral protein genome-linked | | Authors: | Kim, J, Hwang, H, Min, H, Yun, H, Cho, K, Pelton, J.G, Wemmer, D.E, Lee, C. | | Deposit date: | 2014-12-24 | | Release date: | 2015-04-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the porcine sapovirus VPg core reveals a stable three-helical bundle with a conserved surface patch.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
4GKF
 
 | | Crystal structure and characterization of Cmr5 protein from Pyrococcus furiosus | | Descriptor: | CRISPR system Cmr subunit Cmr5 | | Authors: | Park, J, Sun, J, Park, S, Hwang, H, Park, M, Shin, M.S. | | Deposit date: | 2012-08-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Cmr5 from Pyrococcus furiosus and its functional implications
Febs Lett., 587, 2013
|
|
3QBQ
 
 | | Crystal structure of extracellular domains of mouse RANK-RANKL complex | | Descriptor: | Tumor necrosis factor ligand superfamily member 11, Tumor necrosis factor receptor superfamily member 11A | | Authors: | Ta, H.M, Nguyen, G.T.T, Jin, H.M, Choi, J.K, Park, H, Kim, N.S, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2011-01-13 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based development of a receptor activator of nuclear factor-kappaB ligand (RANKL) inhibitor peptide and molecular basis for osteopetrosis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M4W
 
 | | Structural basis for the negative regulation of bacterial stress response by RseB | | Descriptor: | Sigma-E factor negative regulatory protein, Sigma-E factor regulatory protein rseB, ZINC ION | | Authors: | Kim, D.Y, Kwon, E, Choi, J.K, Hwang, H.-Y, Kim, K.K. | | Deposit date: | 2010-03-12 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the negative regulation of bacterial stress response by RseB
Protein Sci., 19, 2010
|
|
5ZOO
 
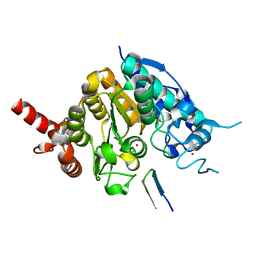 | | Crystal structure of histone deacetylase 4 (HDAC4) in complex with a SMRT corepressor SP1 fragment | | Descriptor: | Histone deacetylase 4, POTASSIUM ION, SMRT corepressor SP1 fragment, ... | | Authors: | Park, S.Y, Hwang, H.J, Kim, J.S. | | Deposit date: | 2018-04-13 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of the specific interaction of SMRT corepressor with histone deacetylase 4.
Nucleic Acids Res., 46, 2018
|
|
5ZOP
 
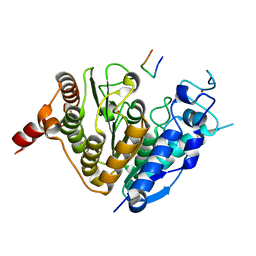 | | Crystal structure of histone deacetylase 4 (HDAC4) in complex with a SMRT corepressor SP2 fragment | | Descriptor: | Histone deacetylase 4, POTASSIUM ION, SMRT corepressor SP2 fragment, ... | | Authors: | Park, S.Y, Hwang, H.J, Kim, J.S. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis of the specific interaction of SMRT corepressor with histone deacetylase 4.
Nucleic Acids Res., 46, 2018
|
|
1L1J
 
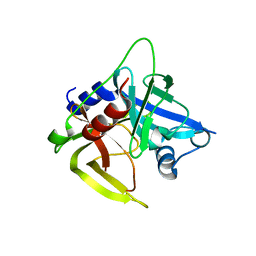 | | Crystal structure of the protease domain of an ATP-independent heat shock protease HtrA | | Descriptor: | heat shock protease HtrA | | Authors: | Kim, D.Y, Kim, D.R, Ha, S.C, Lokanath, N.K, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2002-02-18 | | Release date: | 2003-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Protease Domain of a Heat-shock Protein HtrA from Thermotoga maritima
J.BIOL.CHEM., 278, 2003
|
|
4LFU
 
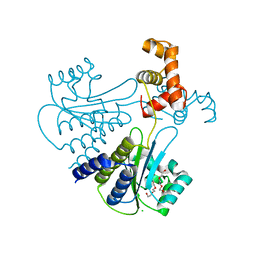 | | Crystal structure of Escherichia coli SdiA in the space group C2 | | Descriptor: | CHLORIDE ION, Regulatory protein SdiA, TETRAETHYLENE GLYCOL | | Authors: | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2013-06-27 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LGW
 
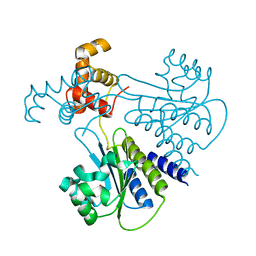 | | Crystal structure of Escherichia coli SdiA in the space group P6522 | | Descriptor: | GLYCEROL, Regulatory protein SdiA | | Authors: | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2013-06-28 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
