4ZLD
 
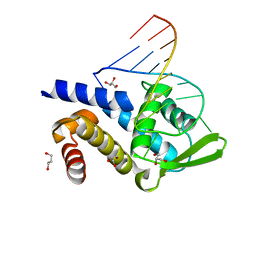 | | Crystal structure of human Roquin-2 ROQ domain in complex with Roquin CDE RNA | | Descriptor: | GLYCEROL, RNA (5'-R(*UP*AP*AP*CP*UP*UP*CP*UP*GP*UP*GP*AP*AP*GP*UP*UP*G)-3'), Roquin-2 | | Authors: | Sakurai, S, Ohto, U, Shimizu, T. | | Deposit date: | 2015-05-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human Roquin-2 and its complex with constitutive-decay element RNA
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7WSI
 
 | |
7VGR
 
 | |
7VGS
 
 | |
5AWA
 
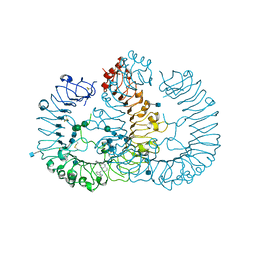 | | Crystal structure of human TLR8 in complex with MB-568 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[[3-(aminomethyl)phenyl]methyl]-3-pentyl-quinolin-2-amine, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2015-07-03 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based Design of Human TLR8-specific Agonists with Augmented Potency and Adjuvanticity
To Be Published
|
|
7VTQ
 
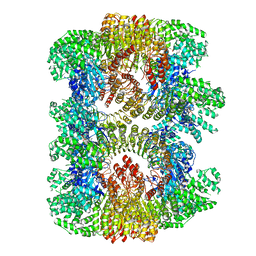 | | Cryo-EM structure of mouse NLRP3 (full-length) dodecamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2021-10-30 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1WDA
 
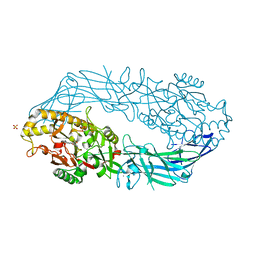 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with benzoyl-L-arginine amide | | Descriptor: | CALCIUM ION, N-[(E)-2-AMINO-1-(3-{[AMINO(IMINO)METHYL]AMINO}PROPYL)-2-HYDROXYVINYL]BENZAMIDE, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
1WD9
 
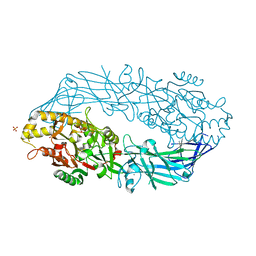 | | Calcium bound form of human peptidylarginine deiminase type4 (PAD4) | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type IV, SULFATE ION | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
1WD8
 
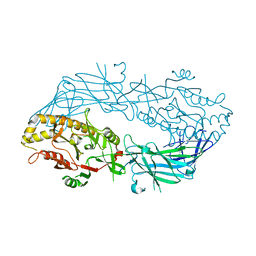 | | Calcium free form of human peptidylarginine deiminase type4 (PAD4) | | Descriptor: | Protein-arginine deiminase type IV | | Authors: | Arita, K, Hashimoto, H, Shimizu, T, Nakashima, K, Yamada, M, Sato, M. | | Deposit date: | 2004-05-12 | | Release date: | 2004-07-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Ca(2+)-induced activation of human PAD4
Nat.Struct.Mol.Biol., 11, 2004
|
|
5ZHX
 
 | |
1V9Y
 
 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (ferric form) | | Descriptor: | Heme pas sensor protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Lee, D.S, Watanabe, M, Sagami, I, Mikami, B, Raman, C.S, Shimizu, T. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
J.Biol.Chem., 279, 2004
|
|
1V9Z
 
 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (Ferrous Form) | | Descriptor: | Heme pas sensor protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Lee, D.S, Watanabe, M, Sagami, I, Mikami, B, Raman, C.S, Shimizu, T. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
J.Biol.Chem., 279, 2004
|
|
1VB6
 
 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (oxygen-bound form) | | Descriptor: | Heme pas sensor protein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Watanabe, M, Sagami, I, Mikami, B, Shimizu, T. | | Deposit date: | 2004-02-24 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of oxygen-bound form of a Heme PAS domain of Ec DOS
To be Published
|
|
1WLF
 
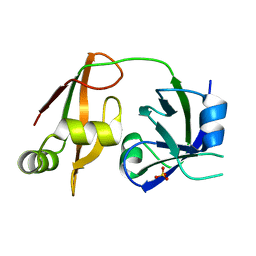 | | Structure of the N-terminal domain of PEX1 AAA-ATPase: Characterization of a putative adaptor-binding domain | | Descriptor: | Peroxisome biogenesis factor 1, SULFATE ION | | Authors: | Shiozawa, K, Maita, N, Tomii, K, Seto, A, Goda, N, Tochio, H, Akiyama, Y, Shimizu, T, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the N-terminal Domain of PEX1 AAA-ATPase: CHARACTERIZATION OF A PUTATIVE ADAPTOR-BINDING DOMAIN
J.Biol.Chem., 279, 2004
|
|
1UHN
 
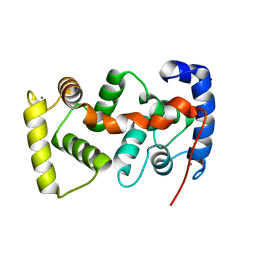 | | The crystal structure of the calcium binding protein AtCBL2 from Arabidopsis thaliana | | Descriptor: | CALCIUM ION, calcineurin B-like protein 2 | | Authors: | Nagae, M, Nozawa, A, Koizumi, N, Sano, H, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2003-07-07 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Novel Calcium-binding Protein AtCBL2 from Arabidopsis thaliana
J.Biol.Chem., 278, 2003
|
|
7VTP
 
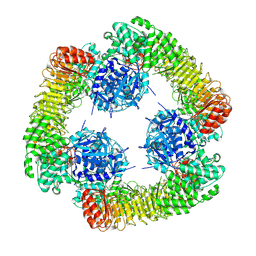 | | Cryo-EM structure of PYD-deleted human NLRP3 hexamer | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2021-10-30 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WBT
 
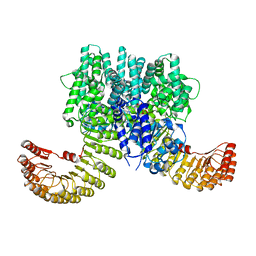 | | Crystal structure of bovine NLRP9 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NACHT, LRR and PYD domains-containing protein 9 | | Authors: | Kamitsukasa, Y, Shimizu, T, Ohto, U. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of NLRP9 reveals a unique C-terminal region with putative regulatory function.
Febs Lett., 596, 2022
|
|
7WBU
 
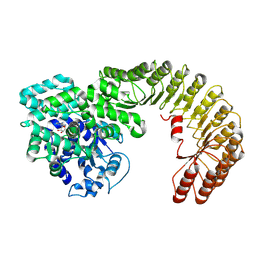 | | Cryo-EM structure of bovine NLRP9 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NACHT, LRR and PYD domains-containing protein 9 | | Authors: | Kamitsukasa, Y, Shimizu, T, Ohto, U. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The structure of NLRP9 reveals a unique C-terminal region with putative regulatory function.
Febs Lett., 596, 2022
|
|
2DM6
 
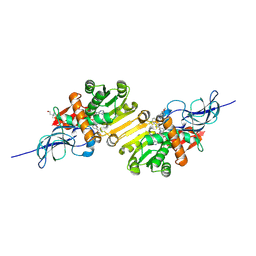 | | Crystal structure of anti-configuration of indomethacin and leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complex | | Descriptor: | INDOMETHACIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent leukotriene B4 12-hydroxydehydrogenase, ... | | Authors: | Hori, T, Ishijima, J, Yokomizo, T, Ago, H, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-20 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of anti-configuration of indomethacin and leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complex reveals the structural basis of broad spectrum indomethacin efficacy
J.Biochem.(Tokyo), 140, 2006
|
|
2COV
 
 | | Crystal structure of CBM31 from beta-1,3-xylanase | | Descriptor: | beta-1,3-xylanase | | Authors: | Hashimoto, H, Tamai, Y, Okazaki, F, Tamaru, Y, Shimizu, T, Araki, T, Sato, M. | | Deposit date: | 2005-05-18 | | Release date: | 2005-09-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The first crystal structure of a family 31 carbohydrate-binding module with affinity to beta-1,3-xylan
Febs Lett., 579, 2005
|
|
2DEX
 
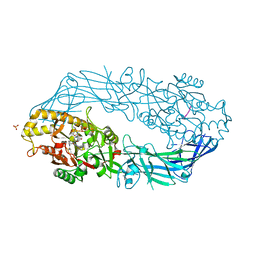 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H3 N-terminal peptide including Arg17 | | Descriptor: | 10-mer peptide from histone H3, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DEW
 
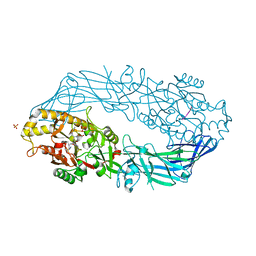 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H3 N-terminal tail including Arg8 | | Descriptor: | 10-mer peptide from histone H3, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DEY
 
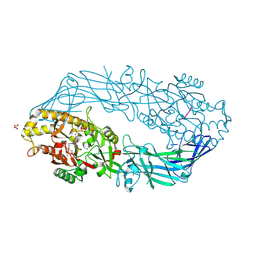 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H4 N-terminal tail including Arg3 | | Descriptor: | 10-mer peptide from histone H4, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2E1N
 
 | |
2CT9
 
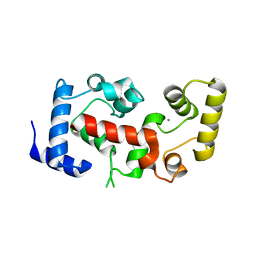 | | The crystal structure of calcineurin B homologous proein 1 (CHP1) | | Descriptor: | CALCIUM ION, Calcium-binding protein p22 | | Authors: | Naoe, Y, Arita, K, Hashimoto, H, Kanazawa, H, Sato, M, Shimizu, T. | | Deposit date: | 2005-05-23 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of calcineurin B homologous protein 1
J.Biol.Chem., 280, 2005
|
|
