7E6J
 
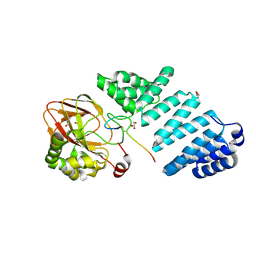 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) H725A in complex with Factor X peptide fragment (39mer-4Ser) | | 分子名称: | Aspartyl/asparaginyl beta-hydroxylase, GLYCEROL, PROPANOIC ACID, ... | | 著者 | Nakashima, Y, Brasnett, A, Brewitz, L, Schofield, C.J. | | 登録日 | 2021-02-22 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Human Oxygenase Variants Employing a Single Protein Fe II Ligand Are Catalytically Active.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DYV
 
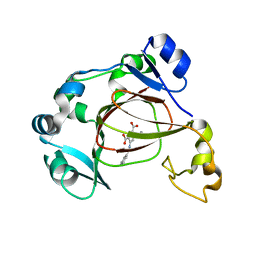 | | Human JMJD5 in complex with MN and 5-(benzylamino)pyridine-2,4-dicarboxylic acid. | | 分子名称: | 5-(benzylamino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | 著者 | Nakashima, Y, Brewitz, L, Schofield, C.J. | | 登録日 | 2021-01-23 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
7DYW
 
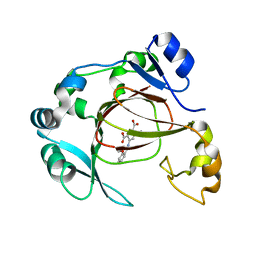 | | Human JMJD5 in complex with MN and 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | 分子名称: | 5-((2-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | 著者 | Nakashima, Y, Brewitz, L, Schofield, C.J. | | 登録日 | 2021-01-23 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
8II0
 
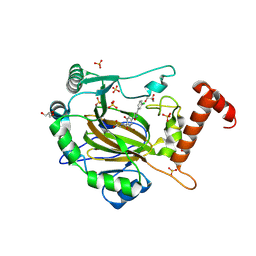 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(3-(3-chlorophenyl)isoxazol-5-yl)-3-hydroxypicolinoyl)glycine | | 分子名称: | 2-[[5-[3-(3-chlorophenyl)-1,2-oxazol-5-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | 著者 | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | 登録日 | 2023-02-24 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-Flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8IHZ
 
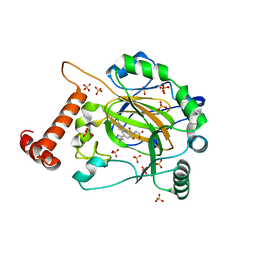 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(1-(3-(4-chlorophenyl)propyl)-1H-1,2,3-triazol-4-yl)-3-hydroxypicolinoyl)glycine | | 分子名称: | 2-[[5-[1-[3-(4-chlorophenyl)propyl]-1,2,3-triazol-4-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | 著者 | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | 登録日 | 2023-02-24 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-Flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8GQ9
 
 | |
8GQB
 
 | |
8GQA
 
 | |
7F8D
 
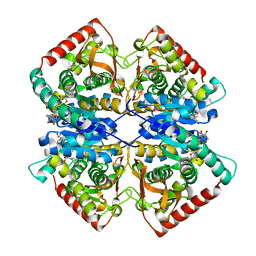 | | Malate Dehydrogenase from Geobacillus stearothermophilus (gs-MDH) G218Y mutant | | 分子名称: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Shimozawa, Y, Himiyama, T, Nakamura, T, Nishiya, Y. | | 登録日 | 2021-07-02 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Increasing loop flexibility affords low-temperature adaptation of a moderate thermophilic malate dehydrogenase from Geobacillus stearothermophilus.
Protein Eng.Des.Sel., 34, 2021
|
|
8ITH
 
 | |
8ITG
 
 | |
4YN5
 
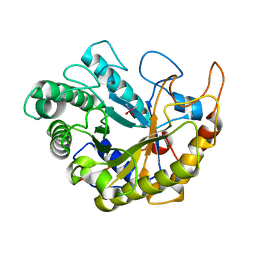 | | Catalytic domain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase | | 分子名称: | CACODYLATE ION, Mannan endo-1,4-beta-mannosidase | | 著者 | Shimane, Y, Ohta, Y, Usami, R, Hatada, Y. | | 登録日 | 2015-03-09 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase
To Be Published
|
|
6LB2
 
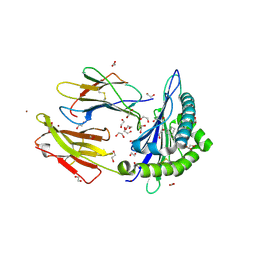 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with mono-acyl glycerol | | 分子名称: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | 著者 | Shima, Y, Morita, D. | | 登録日 | 2019-11-13 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.69380951 Å) | | 主引用文献 | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
7CT4
 
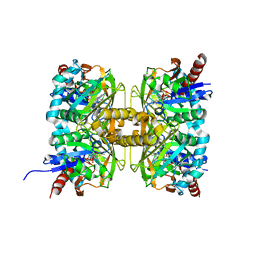 | | Crystal structure of D-amino acid oxidase from Rasamsonia emersonii strain YA | | 分子名称: | D-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Shimekake, Y, Hirato, Y, Okazaki, S, Funabashi, R, Goto, M, Furuichi, T, Suzuki, H, Takahashi, S. | | 登録日 | 2020-08-18 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray structure analysis of a unique D-amino-acid oxidase from the thermophilic fungus Rasamsonia emersonii strain YA.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1IZ0
 
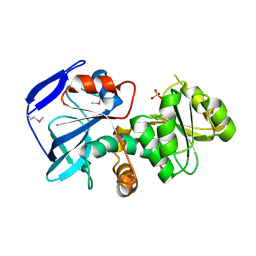 | |
1IYZ
 
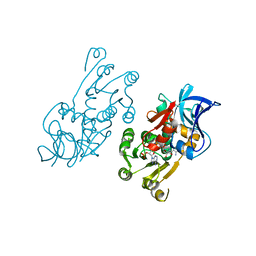 | |
1UJ8
 
 | |
7LLQ
 
 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | 分子名称: | 1-benzyl-4-methoxybenzene, 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, Leukotriene A-4 hydrolase, ... | | 著者 | Lee, K.H, Lee, S.H, Shim, Y.M, Paige, M, Noble, S.M. | | 登録日 | 2021-02-04 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
7KZE
 
 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | 分子名称: | 1-benzyl-4-methoxybenzene, Leukotriene A-4 hydrolase, TRIETHYLENE GLYCOL, ... | | 著者 | Lee, K.H, Shim, Y, Paige, M, Noble, S.M. | | 登録日 | 2020-12-10 | | 公開日 | 2022-06-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
3W08
 
 | | Crystal structure of aldoxime dehydratase | | 分子名称: | Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Hashimoto, H, Nomura, J, Hashimoto, Y, Oinuma, K.I, Wada, K, Hishiki, A, Hara, K, Kobayashi, M. | | 登録日 | 2012-10-24 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of aldoxime dehydratase and its catalytic mechanism involved in carbon-nitrogen triple-bond synthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1QQI
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING AND TRANSACTIVATION DOMAIN OF PHOB FROM ESCHERICHIA COLI | | 分子名称: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | 著者 | Okamura, H, Hanaoka, S, Nagadoi, A, Makino, K, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1999-06-07 | | 公開日 | 2000-06-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.
J.Mol.Biol., 295, 2000
|
|
6ZBO
 
 | | HIF Prolyl Hydroxylase 2 (PHD2/EGLN1) in Complex with 1-(6-morpholinopyrimidin-4-yl)-4-(1H-1,2,3-triazol-1-yl)-1H-pyrazol-5-ol (Molidustat) | | 分子名称: | 2-(6-morpholin-4-ylpyrimidin-4-yl)-4-(1,2,3-triazol-1-yl)pyrazol-3-ol, CHLORIDE ION, Egl nine homolog 1, ... | | 著者 | Figg Jr, W.D, McDonough, M.A, Nakashima, Y, Holt-Martyn, J.P, Schofield, C.J. | | 登録日 | 2020-06-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural Basis of Prolyl Hydroxylase Domain Inhibition by Molidustat.
Chemmedchem, 16, 2021
|
|
6ZBN
 
 | | HIF Prolyl Hydroxylase 2 (PHD2/EGLN1) in complex with tert-butyl 6-(5-hydroxy-4-(1H-1,2,3-triazol-1-yl)-1H-pyrazol-1-yl)nicotinate (IOX4) | | 分子名称: | Egl nine homolog 1, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Figg Jr, W.D, McDonough, M.A, Nakashima, Y, Schofield, C.J. | | 登録日 | 2020-06-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural Basis of Prolyl Hydroxylase Domain Inhibition by Molidustat.
Chemmedchem, 16, 2021
|
|
2MLO
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a Membrane bound form | | 分子名称: | MCP-1 receptor | | 著者 | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | 登録日 | 2014-03-04 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
3VJR
 
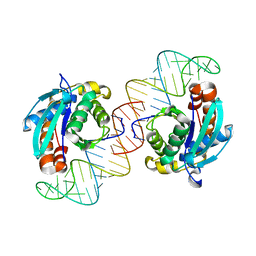 | | Crystal structure of Peptidyl-tRNA hydrolase from Escherichia coli in complex with the CCA-acceptor-T[PSI]C domain of tRNA | | 分子名称: | Peptidyl-tRNA hydrolase, tRNA CCA-acceptor | | 著者 | Ito, K, Murakami, R, Mochizuki, M, Qi, H, Shimizu, Y, Miura, K.I, Ueda, T, Uchiumi, T. | | 登録日 | 2011-10-28 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for the substrate recognition and catalysis of peptidyl-tRNA hydrolase.
Nucleic Acids Res., 40, 2012
|
|
