4Z8M
 
 | | Crystal structure of the MAVS-TRAF6 complex | | Descriptor: | Peptide from Mitochondrial antiviral-signaling protein, TNF receptor-associated factor 6 | | Authors: | Shi, Z.B, Zhou, Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Insights into Mitochondrial Antiviral Signaling Protein (MAVS)-Tumor Necrosis Factor Receptor-associated Factor 6 (TRAF6) Signaling
J.Biol.Chem., 290, 2015
|
|
6WG6
 
 | |
6WG3
 
 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex | | Descriptor: | Cohesin subunit SA-1, DNA (51-MER), Double-strand-break repair protein rad21 homolog, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
6WG4
 
 | |
6WGE
 
 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex without STAG1 | | Descriptor: | DNA (43-MER), Double-strand-break repair protein rad21 homolog, MAGNESIUM ION, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
7W1M
 
 | | Cryo-EM structure of human cohesin-CTCF-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cohesin subunit SA-1, ... | | Authors: | Shi, Z.B, Bai, X.C, Yu, H. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | CTCF and R-loops are boundaries of cohesin-mediated DNA looping.
Mol.Cell, 83, 2023
|
|
4FZF
 
 | | Crystal structure of MST4-MO25 complex with DKI | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZD
 
 | | Crystal structure of MST4-MO25 complex with WSF motif | | Descriptor: | C-terminal peptide from Serine/threonine-protein kinase MST4, Calcium-binding protein 39, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZA
 
 | | Crystal structure of MST4-MO25 complex | | Descriptor: | Calcium-binding protein 39, GLYCEROL, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
5XY9
 
 | | Structure of the MST4 and 14-3-3 complex | | Descriptor: | 14-3-3 protein zeta/delta, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2017-07-06 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structure of the MST4 and 14-3-3 complex
To Be Published
|
|
4RV0
 
 | | Crystal structure of TN complex | | Descriptor: | Nuclear protein localization protein 4 homolog, SULFATE ION, Transitional endoplasmic reticulum ATPase TER94 | | Authors: | Hao, Q, Jiao, S, Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2014-11-23 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of TN complex
To be Published
|
|
8Y3M
 
 | | Cryo-EM structure of DSR2-DSAD1 complex (cross-linked) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y3W
 
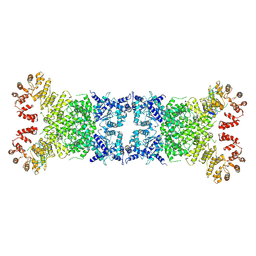 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (same side) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y13
 
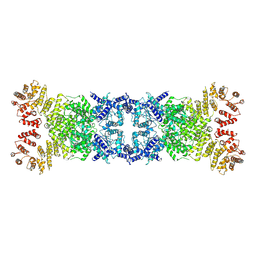 | | Cryo-EM structure of anti-phage defense associated DSR2 tetramer (H171A) | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Li, F.X, Shi, Z.B, Wang, R.W, Xu, Q, Yang, R, Wu, Z.X. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y3Y
 
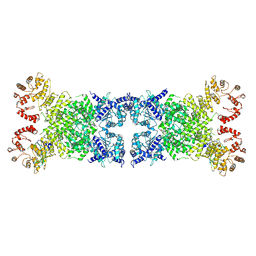 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (opposite side) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y34
 
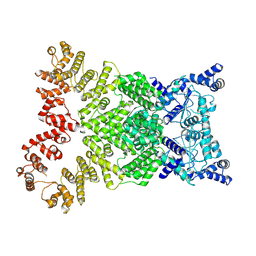 | | Cryo-EM structure of anti-phage defense associated DSR2 (H171A) (map2) | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-28 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
5GZB
 
 | | Crystal Structure of Transcription Factor TEAD4 in Complex with M-CAT DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*GP*GP*AP*AP*TP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*AP*TP*TP*CP*CP*TP*CP*TP*C)-3'), GLYCEROL, ... | | Authors: | He, F, Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-04-19 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | DNA-binding mechanism of the Hippo pathway transcription factor TEAD4
Oncogene, 36, 2017
|
|
4NT4
 
 | | Crystal structure of the kinase domain of Gilgamesh isoform I from Drosophila melanogaster | | Descriptor: | GLYCEROL, Gilgamesh, isoform I, ... | | Authors: | Chen, C.C, Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2013-11-30 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of the kinase domain of Gilgamesh from Drosophila melanogaster
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4KZG
 
 | |
4KGB
 
 | |
4HKD
 
 | |
4ZDR
 
 | | Crystal structure of 14-3-3[zeta]-LKB1 fusion protein | | Descriptor: | 14-3-3 protein zeta/delta,GGSGGS linker,Serine/threonine-protein kinase STK11, GLYCEROL, PROPANE, ... | | Authors: | Ding, S, Shi, Z.B. | | Deposit date: | 2015-04-18 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structure of the 14-3-3 zeta-LKB1 fusion protein provides insight into a novel ligand-binding mode of 14-3-3.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4GEH
 
 | |
4FI9
 
 | | Structure of human SUN-KASH complex | | Descriptor: | Nesprin-2, SUN domain-containing protein 2 | | Authors: | Wang, W.J, Shi, Z.B. | | Deposit date: | 2012-06-08 | | Release date: | 2012-07-18 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into SUN-KASH complexes across the nuclear envelope.
Cell Res., 22, 2012
|
|
4HK1
 
 | |
