6KLH
 
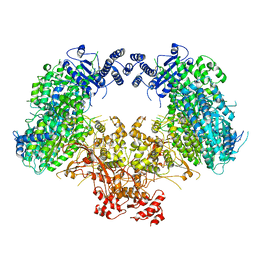 | | Dimeric structure of Machupo virus polymerase bound to vRNA promoter | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*GP*CP*CP*UP*AP*GP*GP*AP*UP*CP*CP*AP*CP*UP*GP*UP*GP*CP*G)-3'), RNA-directed RNA polymerase L, ... | | Authors: | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | Deposit date: | 2019-07-30 | | Release date: | 2020-03-18 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
6KLE
 
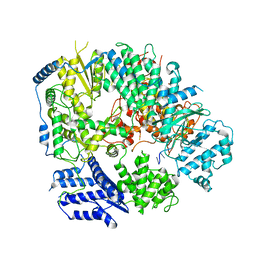 | | Monomeric structure of Machupo virus polymerase bound to vRNA promoter | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*GP*CP*CP*UP*AP*GP*GP*AP*UP*CP*CP*AP*CP*UP*GP*UP*GP*CP*G)-3'), RNA-directed RNA polymerase L, ... | | Authors: | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | Deposit date: | 2019-07-30 | | Release date: | 2020-03-18 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
6KLD
 
 | | Structure of apo Machupo virus polymerase | | Descriptor: | MANGANESE (II) ION, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | Deposit date: | 2019-07-30 | | Release date: | 2020-03-18 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
1DU9
 
 | | SOLUTION STRUCTURE OF BMP02, A NATURAL SCORPION TOXIN WHICH BLOCKS APAMIN-SENSITIVE CALCIUM-ACTIVATED POTASSIUM CHANNELS, 25 STRUCTURES | | Descriptor: | BMP02 NEUROTOXIN | | Authors: | Xu, Y, Wu, J, Pei, J, Shi, Y, Ji, Y, Tong, Q. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-04 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmP02, a new potassium channel blocker from the venom of the Chinese scorpion Buthus martensi Karsch.
Biochemistry, 39, 2000
|
|
3IA3
 
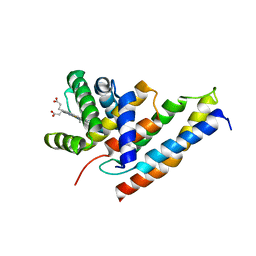 | |
3J8H
 
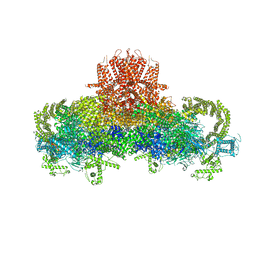 | | Structure of the rabbit ryanodine receptor RyR1 in complex with FKBP12 at 3.8 Angstrom resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Yan, Z, Bai, X, Yan, C, Wu, J, Scheres, S.H.W, Shi, Y, Yan, N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution.
Nature, 517, 2015
|
|
4M69
 
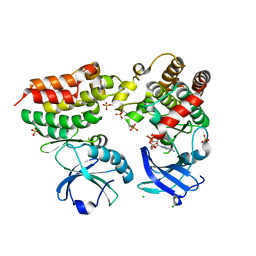 | | Crystal structure of the mouse RIP3-MLKL complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mixed lineage kinase domain-like protein, ... | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
3JB9
 
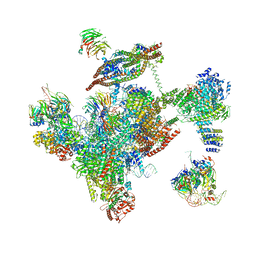 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
3JCM
 
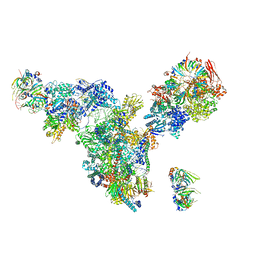 | | Cryo-EM structure of the spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Wan, R, Yan, C, Bai, R, Wang, L, Huang, M, Wong, C.C, Shi, Y. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis
Science, 351, 2016
|
|
3K7V
 
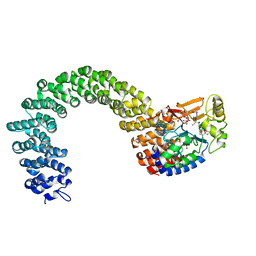 | | Protein phosphatase 2A core complex bound to dinophysistoxin-1 | | Descriptor: | (2R)-3-[(2S,5R,6R,8S)-8-{(1R,2E)-3-[(2R,4a'R,5R,6'S,8'R,8a'S)-6'-{(1S,3S)-3-[(2S,3R,6R,11R)-3,11-dimethyl-1,7-dioxaspiro[5.5]undec-2-yl]-1-hydroxybutyl}-8'-hydroxy-7'-methylideneoctahydro-3H,3'H-spiro[furan-2,2'-pyrano[3,2-b]pyran]-5-yl]-1-methylprop-2-en-1-yl}-5-hydroxy-10-methyl-1,7-dioxaspiro[5.5]undec-10-en-2-yl]-2-hydroxy-2-methylpropanoic acid, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Jeffrey, P.D, Huhn, J, Shi, Y. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A structural basis for the reduced toxicity of dinophysistoxin-2.
Chem.Res.Toxicol., 22, 2009
|
|
3KV9
 
 | | Structure of KIAA1718 Jumonji domain | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, OXYGEN MOLECULE | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4LNR
 
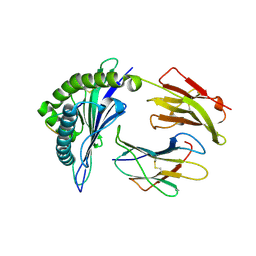 | | The structure of HLA-B*35:01 in complex with the peptide (RPQVPLRPMTY) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Cheng, H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-12 | | Release date: | 2014-07-23 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide-dependent conformational fluctuation determines the stability of the human leukocyte antigen class I complex.
J.Biol.Chem., 289, 2014
|
|
4LL9
 
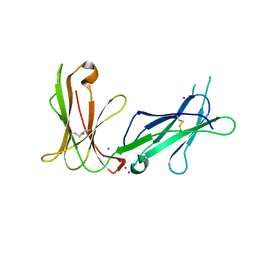 | | Crystal structure of D3D4 domain of the LILRB1 molecule | | Descriptor: | IODIDE ION, Leukocyte immunoglobulin-like receptor subfamily B member 1 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
3K7W
 
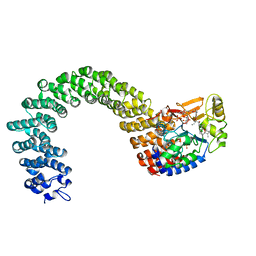 | | Protein phosphatase 2A core complex bound to dinophysistoxin-2 | | Descriptor: | (2R)-2-hydroxy-3-[(2S,5R,6R,8S)-5-hydroxy-8-{(1R,2E)-3-[(2R,4a'R,5R,6'S,8'R,8a'S)-8'-hydroxy-6'-{(1S,3S)-1-hydroxy-3-[( 2S,6R,11S)-11-methyl-1,7-dioxaspiro[5.5]undec-2-yl]butyl}-7'-methylideneoctahydro-3H,3'H-spiro[furan-2,2'-pyrano[3,2-b]p yran]-5-yl]-1-methylprop-2-en-1-yl}-10-methyl-1,7-dioxaspiro[5.5]undec-10-en-2-yl]-2-methylpropanoic acid, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Jeffrey, P.D, Huhn, J, Shi, Y. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | A structural basis for the reduced toxicity of dinophysistoxin-2.
Chem.Res.Toxicol., 22, 2009
|
|
3KVB
 
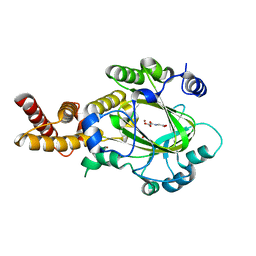 | | Structure of KIAA1718 Jumonji domain in complex with N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4M68
 
 | | Crystal structure of the mouse MLKL kinase-like domain | | Descriptor: | GLYCEROL, Mixed lineage kinase domain-like protein | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
4M66
 
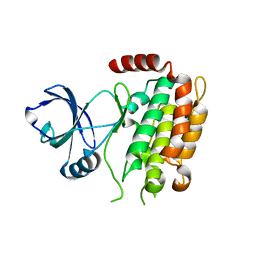 | | Crystal structure of the mouse RIP3 kinase domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
4LLA
 
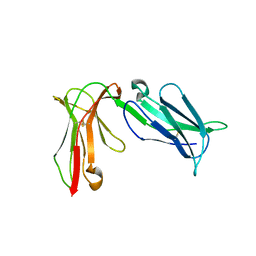 | | Crystal structure of D3D4 domain of the LILRB2 molecule | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 2 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
8I0V
 
 | | The cryo-EM structure of human post-Bact complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0P
 
 | | The cryo-EM structure of human pre-Bact complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0W
 
 | | The cryo-EM structure of human C complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Coiled-coil domain-containing protein 94, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0R
 
 | | The cryo-EM structure of human Bact-I complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0U
 
 | | The cryo-EM structure of human Bact-IV complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0T
 
 | | The cryo-EM structure of human Bact-III complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0S
 
 | | The cryo-EM structure of human Bact-II complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
