7EVN
 
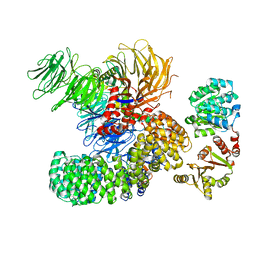 | | The cryo-EM structure of the DDX42-SF3b complex | | Descriptor: | ATP-dependent RNA helicase DDX42, PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome
Nat.Struct.Mol.Biol., 2024
|
|
1F0K
 
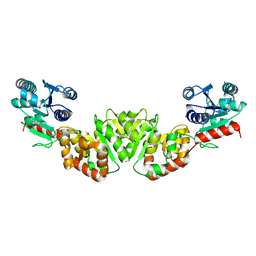 | | THE 1.9 ANGSTROM CRYSTAL STRUCTURE OF E. COLI MURG | | Descriptor: | SULFATE ION, UDP-N-ACETYLGLUCOSAMINE-N-ACETYLMURAMYL-(PENTAPEPTIDE) PYROPHOSPHORYL-UNDECAPRENOL N-ACETYLGLUCOSAMINE TRANSFERASE | | Authors: | Ha, S, Walker, D, Shi, Y, Walker, S. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis.
Protein Sci., 9, 2000
|
|
2OSG
 
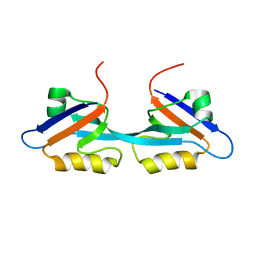 | | Solution Structure and Binding Property of the Domain-swapped Dimer of ZO2PDZ2 | | Descriptor: | Tight junction protein ZO-2 | | Authors: | Wu, J.W, Yang, Y.S, Zhang, J.H, Ji, P, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2007-02-05 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Domain-swapped Dimerization of the Second PDZ Domain of ZO2 May Provide a Structural Basis for the Polymerization of Claudins
J.Biol.Chem., 282, 2007
|
|
7DVQ
 
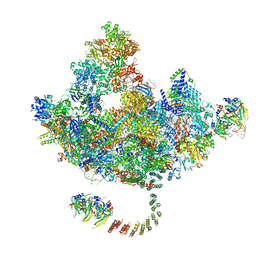 | | Cryo-EM Structure of the Activated Human Minor Spliceosome (minor Bact Complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'-O-[(S)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(methoxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]guanosine, Armadillo repeat-containing protein 7, ... | | Authors: | Bai, R, Wan, R, Wang, L, Xu, K, Zhang, Q, Lei, J, Shi, Y. | | Deposit date: | 2021-01-14 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the activated human minor spliceosome.
Science, 371, 2021
|
|
2PKG
 
 | |
7DCO
 
 | | Cryo-EM structure of the activated spliceosome (Bact complex) at an atomic resolution of 2.5 angstrom | | Descriptor: | BJ4_G0014900.mRNA.1.CDS.1, BJ4_G0027490.mRNA.1.CDS.1, BJ4_G0037700.mRNA.1.CDS.1, ... | | Authors: | Bai, R, Wan, R, Yan, C, Qi, J, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7DCR
 
 | | cryo-EM structure of the DEAH-box helicase Prp2 in complex with its coactivator Spp2 | | Descriptor: | PRP2 isoform 1, Pre-mRNA-splicing factor SPP2 | | Authors: | Bai, R, Wan, R, Yan, C, Jia, Q, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
1P1E
 
 | |
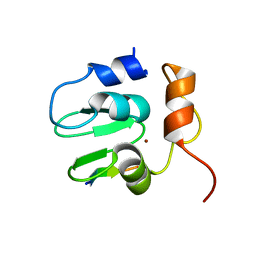
1I51
 
 | | CRYSTAL STRUCTURE OF CASPASE-7 COMPLEXED WITH XIAP | | Descriptor: | CASPASE-7 SUBUNIT P11, CASPASE-7 SUBUNIT P20, X-LINKED INHIBITOR OF APOPTOSIS PROTEIN | | Authors: | Chai, J, Shi, Y. | | Deposit date: | 2001-02-23 | | Release date: | 2002-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of caspase-7 inhibition by XIAP.
Cell(Cambridge,Mass.), 104, 2001
|
|
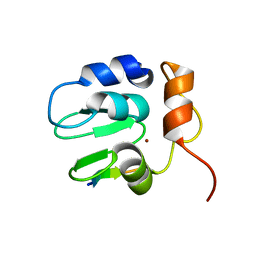
1SE0
 
 | | Crystal structure of DIAP1 BIR1 bound to a Grim peptide | | Descriptor: | Apoptosis 1 inhibitor, Cell death protein Grim, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SDZ
 
 | | Crystal structure of DIAP1 BIR1 bound to a Reaper peptide | | Descriptor: | Apoptosis 1 inhibitor, Reaper, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
7DD3
 
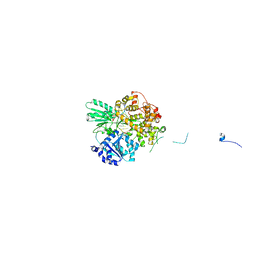 | | Cryo-EM structure of the pre-mRNA-loaded DEAH-box ATPase/helicase Prp2 in complex with Spp2 | | Descriptor: | PRP2 isoform 1, Pre-mRNA-splicing factor SPP2, pre-mRNA | | Authors: | Bai, R, Wan, R, Yan, C, Qi, J, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7DCP
 
 | | cryo-EM structure of the DEAH-box helicase Prp2 and coactivator Spp2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PRP2 isoform 1, ... | | Authors: | Bai, R, Wan, R, Yan, C, Jia, Q, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7EP7
 
 | | The complex structure of Gpsm2 and Whirlin | | Descriptor: | G-protein-signaling modulator 2, Whirlin | | Authors: | Lin, L, Shi, Y, Wang, C, Zhu, J. | | Deposit date: | 2021-04-26 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Promotion of row 1-specific tip complex condensates by Gpsm2-G alpha i provides insights into row identity of the tallest stereocilia.
Sci Adv, 8, 2022
|
|
1JD5
 
 | | Crystal Structure of DIAP1-BIR2/GRIM | | Descriptor: | APOPTOSIS 1 INHIBITOR, ZINC ION, cell death protein GRIM | | Authors: | Wu, J.W, Cocina, A.E, Chai, J, Hay, B.A, Shi, Y. | | Deposit date: | 2001-06-12 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Mol.Cell, 8, 2001
|
|
1JD6
 
 | | Crystal Structure of DIAP1-BIR2/Hid Complex | | Descriptor: | APOPTOSIS 1 INHIBITOR, ZINC ION, head involution defective protein | | Authors: | Wu, J.W, Cocina, A.E, Chai, J, Hay, B.A, Shi, Y. | | Deposit date: | 2001-06-12 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Mol.Cell, 8, 2001
|
|
1JD4
 
 | | Crystal Structure of DIAP1-BIR2 | | Descriptor: | APOPTOSIS 1 INHIBITOR, ZINC ION | | Authors: | Wu, J.W, Cocina, A.E, Chai, J, Hay, B.A, Shi, Y. | | Deposit date: | 2001-06-12 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Mol.Cell, 8, 2001
|
|
1G73
 
 | | CRYSTAL STRUCTURE OF SMAC BOUND TO XIAP-BIR3 DOMAIN | | Descriptor: | INHIBITORS OF APOPTOSIS-LIKE PROTEIN ILP, SECOND MITOCHONDRIA-DERIVED ACTIVATOR OF CASPASES, ZINC ION | | Authors: | Wu, G, Chai, J, Suber, T.L, Wu, J.-W, Shi, Y. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of IAP recognition by Smac/DIABLO.
Nature, 408, 2000
|
|
1P1D
 
 | |
2P1L
 
 | |
7DCQ
 
 | | cryo-EM structure of the DEAH-box helicase Prp2 | | Descriptor: | PRP2 isoform 1 | | Authors: | Bai, R, Wan, R, Yan, C, Jia, Q, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7BCQ
 
 | | ASCT2 in the presence of the inhibitor Lc-BPE (position "up") in the outward-open conformation. | | Descriptor: | 4-(4-phenylphenyl)carbonyloxypyrrolidine-2-carboxylic acid, Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BCT
 
 | | ASCT2 in the presence of the inhibitor ERA-21 in the outward-open conformation. | | Descriptor: | Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D8X
 
 | | CryoEM structure of human gamma-secretase in complex with E2012 and L685458 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[(1S)-1-(4-fluorophenyl)ethyl]-3-[[3-methoxy-4-(4-methylimidazol-1-yl)phenyl]methylidene]piperidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
7C9I
 
 | | Human gamma-secretase in complex with small molecule L-685,458 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
