4YYA
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with avian receptor analog 3'SLNLN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | 著者 | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | 登録日 | 2015-03-23 | | 公開日 | 2016-04-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.595 Å) | | 主引用文献 | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
8A9L
 
 | | Cryo-EM structure of alpha-synuclein filaments from Parkinson's disease and dementia with Lewy bodies | | 分子名称: | Alpha-synuclein, Unknown fragment | | 著者 | Yang, Y, Shi, Y, Schweighauser, M, Zhang, X.J, Kotecha, A, Murzin, A.G, Garringer, H.J, Cullinane, P, Saito, Y, Foroud, T, Warner, T.T, Hasegawa, K, Vidal, R, Murayama, S, Revesz, T, Ghetti, B, Hasegawa, M, Lashley, T, Scheres, H.W.S, Goedert, M. | | 登録日 | 2022-06-28 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-11-09 | | 実験手法 | ELECTRON MICROSCOPY (2.16 Å) | | 主引用文献 | Structures of alpha-synuclein filaments from human brains with Lewy pathology.
Nature, 610, 2022
|
|
5JHL
 
 | | Crystal structure of zika virus envelope protein in complex with a flavivirus broadly-protective antibody | | 分子名称: | Antibody Heavy chain, antibody Light chain, envelope protein | | 著者 | Dai, L, Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2016-04-21 | | 公開日 | 2016-05-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody.
Cell Host Microbe, 19, 2016
|
|
4QY0
 
 | | Structure of H10 from human-infecting H10N8 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | 著者 | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | 登録日 | 2014-07-23 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
7TA8
 
 | | NMR structure of crosslinked cyclophilin A | | 分子名称: | Peptidyl-prolyl cis-trans isomerase A | | 著者 | Lu, M, Toptygin, D, Xiang, Y, Shi, Y, Schwieters, C.D, Lipinski, E.C, Ahn, J, Byeon, I.-J.L, Gronenborn, A.M. | | 登録日 | 2021-12-20 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Magic of Linking Rings: Discovery of a Unique Photoinduced Fluorescent Protein Crosslink.
J.Am.Chem.Soc., 144, 2022
|
|
4YYB
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with human receptor analog 6'SLNLN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | 著者 | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | 登録日 | 2015-03-23 | | 公開日 | 2016-04-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.609 Å) | | 主引用文献 | Structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) at 2.6 Angstroms resolution
To Be Published
|
|
4QY2
 
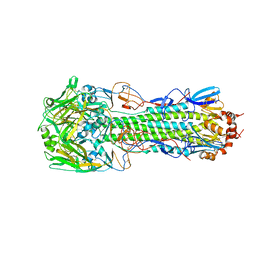 | | Structure of H10 from human-infecting H10N8 virus in complex with human receptor analog | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, hemagglutinin | | 著者 | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | 登録日 | 2014-07-23 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.399 Å) | | 主引用文献 | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
5JCP
 
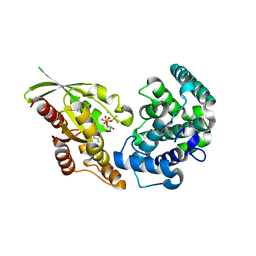 | | RhoGAP domain of ARAP3 in complex with RhoA in the transition state | | 分子名称: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3,Linker,Transforming protein RhoA, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Bao, H, Li, F, Wang, C, Wang, N, Jiang, Y, Tang, Y, Wu, J, Shi, Y. | | 登録日 | 2016-04-15 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for the Specific Recognition of RhoA by the Dual GTPase-activating Protein ARAP3
J.Biol.Chem., 291, 2016
|
|
7N9A
 
 | |
5JD0
 
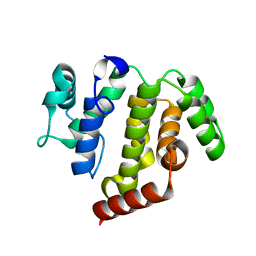 | | crystal structure of ARAP3 RhoGAP domain | | 分子名称: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | 著者 | Bao, H, Li, F, Wu, J, Shi, Y. | | 登録日 | 2016-04-15 | | 公開日 | 2016-06-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis for the Specific Recognition of RhoA by the Dual GTPase-activating Protein ARAP3
J.Biol.Chem., 291, 2016
|
|
4Y6K
 
 | | Complex structure of presenilin homologue PSH bound to an inhibitor | | 分子名称: | N-{(2R,4S,5S)-2-benzyl-5-[(tert-butoxycarbonyl)amino]-4-hydroxy-6-phenylhexanoyl}-L-leucyl-L-phenylalaninamide, Uncharacterized protein PSH | | 著者 | Dang, S, Wu, S, Wang, J, Shi, Y. | | 登録日 | 2015-02-13 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.855 Å) | | 主引用文献 | Cleavage of amyloid precursor protein by an archaeal presenilin homologue PSH
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4R8W
 
 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a neutralizing antibody CT149 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of neutralizing antibody CT149, Hemagglutinin, ... | | 著者 | Wu, Y, Shi, Y, Qi, J, Gao, G.F. | | 登録日 | 2014-09-03 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.795 Å) | | 主引用文献 | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus
Nat Commun, 6, 2015
|
|
4YY0
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2 | | 著者 | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | 登録日 | 2015-03-23 | | 公開日 | 2016-03-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4Q9V
 
 | | Crystal structure of TIPE3 | | 分子名称: | CHLORIDE ION, SULFATE ION, Tumor necrosis factor alpha-induced protein 8-like protein 3 | | 著者 | Wu, J, Zhang, X, Chen, Y.H, Shi, Y. | | 登録日 | 2014-05-02 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer.
Cancer Cell, 26, 2014
|
|
4R12
 
 | | Crystal structure of the gamma-secretase component Nicastrin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Xie, T, Yan, C, Zhou, R, Zhao, Y, Sun, L, Yang, G, Lu, P, Ma, D, Shi, Y. | | 登録日 | 2014-08-03 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the gamma-secretase component nicastrin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QVD
 
 | | E.coli Hfq in complex with RNA Ads | | 分子名称: | RNA (5'-R(*AP*AP*CP*UP*AP*AP*A)-3'), RNA-binding protein Hfq | | 著者 | Wang, L.J, Wang, W.W, Li, F.D, Wu, J.H, Gong, Q.G, Shi, Y.Y. | | 登録日 | 2014-07-14 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.972 Å) | | 主引用文献 | Structural insights into the recognition of the internal A-rich linker from OxyS sRNA by Escherichia coli Hfq
Nucleic Acids Res., 43, 2015
|
|
4QVC
 
 | | E.coli Hfq in complex with RNA Aus | | 分子名称: | RNA (5'-R(*AP*U*AP*AP*CP*UP*A)-3'), RNA-binding protein Hfq | | 著者 | Wang, L.J, Wang, W.W, Li, F.D, Wu, J.H, Gong, Q.G, Shi, Y.Y. | | 登録日 | 2014-07-14 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural insights into the recognition of the internal A-rich linker from OxyS sRNA by Escherichia coli Hfq
Nucleic Acids Res., 43, 2015
|
|
5DNO
 
 | | Crystal structure of Mmi1 YTH domain complex with RNA | | 分子名称: | RNA (5'-R(*CP*UP*UP*AP*AP*AP*C)-3'), YTH domain-containing protein mmi1 | | 著者 | Wang, C.Y, Zhu, Y.W, Wu, J.H, Shi, Y.Y. | | 登録日 | 2015-09-10 | | 公開日 | 2016-04-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A novel RNA-binding mode of the YTH domain reveals the mechanism for recognition of determinant of selective removal by Mmi1
Nucleic Acids Res., 44, 2016
|
|
5DNP
 
 | | Crystal structure of Mmi1 YTH domain | | 分子名称: | PHOSPHATE ION, YTH domain-containing protein mmi1 | | 著者 | Wang, C.Y, Zhu, Y.W, Shi, Y.Y, Wu, J.H. | | 登録日 | 2015-09-10 | | 公開日 | 2016-04-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A novel RNA-binding mode of the YTH domain reveals the mechanism for recognition of determinant of selective removal by Mmi1
Nucleic Acids Res., 44, 2016
|
|
1ECU
 
 | | SOLUTION STRUCTURE OF E2F BINDING DNA FRAGMENT GCGCGAAAC-T-GTTTCGCGC | | 分子名称: | DNA (5'-D(*GP*CP*GP*CP*GP*AP*AP*AP*CP*TP*GP*TP*TP*TP*CP*GP*CP*GP*C)-3') | | 著者 | Wu, J.H, Chang, C, Pei, J.M, Xiao, Q, Shi, Y.Y. | | 登録日 | 2000-01-26 | | 公開日 | 2000-02-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of E2F binding DNA fragment GCGCGAAAC-T-GTTTCGCGC studied by Molecular Dynamics Simulation and Two Dimensional NMR experiment
to be published, 2000
|
|
1HW5
 
 | | THE CAP/CRP VARIANT T127L/S128A | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR | | 著者 | Chu, S.Y, Tordova, M, Gilliland, G.L, Gorshkova, I, Shi, Y. | | 登録日 | 2001-01-09 | | 公開日 | 2001-01-17 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | The structure of the T127L/S128A mutant of cAMP receptor protein facilitates promoter site binding
J.Biol.Chem., 276, 2001
|
|
5JNX
 
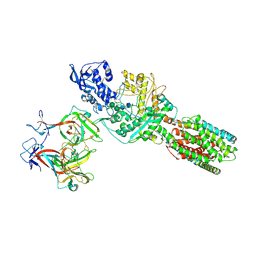 | | The 6.6 A cryo-EM structure of the full-length human NPC1 in complex with the cleaved glycoprotein of Ebola virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, ... | | 著者 | Gong, X, Qian, H.W, Zhou, X.H, Wu, J.P, Wan, T, Shi, Y, Gao, F, Zhou, Q, Yan, N. | | 登録日 | 2016-05-01 | | 公開日 | 2016-06-15 | | 最終更新日 | 2020-08-05 | | 実験手法 | ELECTRON MICROSCOPY (6.56 Å) | | 主引用文献 | Structural Insights into the Niemann-Pick C1 (NPC1)-Mediated Cholesterol Transfer and Ebola Infection
Cell, 165, 2016
|
|
3PXG
 
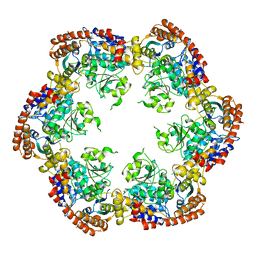 | | Structure of MecA121 and ClpC1-485 complex | | 分子名称: | Adapter protein mecA 1, Negative regulator of genetic competence ClpC/MecB | | 著者 | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | 登録日 | 2010-12-09 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.654 Å) | | 主引用文献 | Structure and mechanism of the hexameric MecA-ClpC molecular machine.
Nature, 471, 2011
|
|
3DW8
 
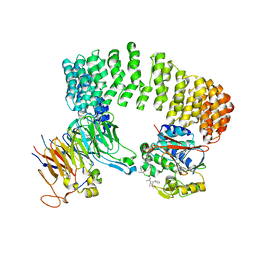 | | Structure of a Protein Phosphatase 2A Holoenzyme with B55 subunit | | 分子名称: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | 著者 | Xu, Y, Chen, Y, Zhang, P, Jeffrey, P.D, Shi, Y. | | 登録日 | 2008-07-21 | | 公開日 | 2008-10-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation.
Mol.Cell, 31, 2008
|
|
4YDE
 
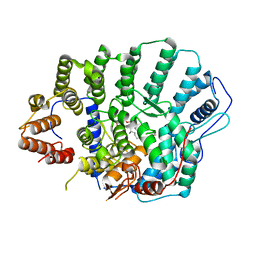 | | CRYSTAL STRUCTURE OF CANDIDA ALBICANS PROTEIN FARNESYLTRANSFERASE BINARY COMPLEX WITH THE ISOPRENOID FARNESYLDIPHOSPHATE | | 分子名称: | (3R,7S)-3,7,11-trimethyldodecyl trihydrogen diphosphate, 1,2-ETHANEDIOL, Protein farnesyltransferase/geranylgeranyltransferase type-1 Subunit beta, ... | | 著者 | Kumar, S, Mabanglo, M.F, Hast, M.A, Shi, Y, Beese, L.S. | | 登録日 | 2015-02-22 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF CANDIDA ALBICANS PROTEIN FARNESYLTRANSFERASE BINARY COMPLEX WITH THE ISOPRENOID FARNESYLDIPHOSPHATE
To Be Published
|
|
