4UIS
 
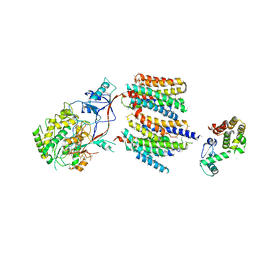 | | The cryoEM structure of human gamma-Secretase complex | | 分子名称: | GAMMA-SECRETASE, LYSOZYME | | 著者 | Sun, L, Zhao, L, Yang, G, Yan, C, Zhou, R, Zhou, X, Xie, T, Zhao, Y, Wu, S, Li, X, Shi, Y. | | 登録日 | 2015-04-03 | | 公開日 | 2015-06-10 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural Basis of Human Gamma-Secretase Assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5E64
 
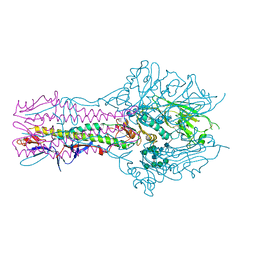 | | Hemagglutinin-esterase-fusion protein structure of influenza D virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, ... | | 著者 | Song, H, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2015-10-09 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | 著者 | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | 登録日 | 2012-08-29 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
3LQR
 
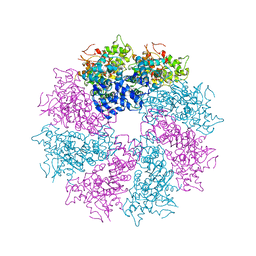 | | Structure of CED-4:CED-3 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | 著者 | Qi, S, Pang, Y, Shi, Y, Yan, N. | | 登録日 | 2010-02-09 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.896 Å) | | 主引用文献 | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
5WVE
 
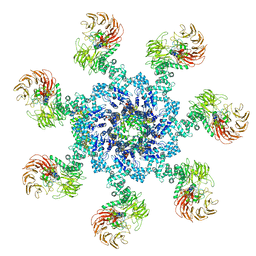 | | Apaf-1-Caspase-9 holoenzyme | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Caspase, ... | | 著者 | Li, Y, Zhou, M, Hu, Q, Shi, Y. | | 登録日 | 2016-12-24 | | 公開日 | 2017-02-08 | | 最終更新日 | 2017-03-01 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ELH
 
 | | Crystal structure of mouse Unkempt zinc fingers 1-3 (ZnF1-3), bound to RNA | | 分子名称: | RING finger protein unkempt homolog, RNA (5'-R(*UP*UP*AP*UP*U)-3'), SULFATE ION, ... | | 著者 | Teplova, M, Murn, J, Zarnack, K, Shi, Y, Patel, D.J. | | 登録日 | 2015-11-04 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Recognition of distinct RNA motifs by the clustered CCCH zinc fingers of neuronal protein Unkempt.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8JOL
 
 | | cryo-EM structure of the CED-4/CED-3 holoenzyme | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 3, Cell death protein 4, ... | | 著者 | Li, Y, Tian, L, Zhang, Y, Shi, Y. | | 登録日 | 2023-06-07 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into CED-3 activation.
Life Sci Alliance, 6, 2023
|
|
5WZJ
 
 | | Structure of APUM23-GGAUUUGACGG | | 分子名称: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*UP*UP*UP*GP*AP*CP*GP*G)-3') | | 著者 | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | 登録日 | 2017-01-18 | | 公開日 | 2017-09-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | 著者 | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | 登録日 | 2016-12-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
5WZH
 
 | | Structure of APUM23-GGAAUUGACGG | | 分子名称: | Pumilio homolog 23, RNA (5'-R(*GP*GP*AP*AP*UP*UP*GP*AP*CP*GP*G)-3') | | 著者 | Bao, H, Wang, N, Wang, C, Jiang, Y, Wu, J, Shi, Y. | | 登録日 | 2017-01-18 | | 公開日 | 2017-09-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.509 Å) | | 主引用文献 | Structural basis for the specific recognition of 18S rRNA by APUM23.
Nucleic Acids Res., 45, 2017
|
|
4GG4
 
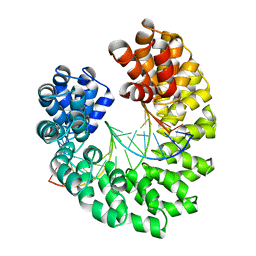 | | Crystal structure of the TAL effector dHax3 bound to specific DNA-RNA hybrid | | 分子名称: | DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3, RNA (5'-R(*AP*GP*AP*GP*AP*GP*AP*UP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3') | | 著者 | Yin, P, Deng, D, Yan, C.Y, Pan, X.J, Yan, N, Shi, Y.G. | | 登録日 | 2012-08-05 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Specific DNA-RNA hybrid recognition by TAL effectors
Cell Rep, 2, 2012
|
|
5WZ1
 
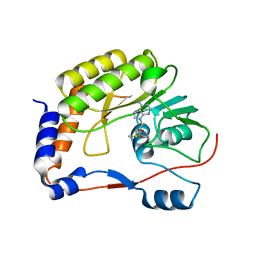 | | Crystal structure of Zika virus NS5 methyltransferase bound to S-adenosyl-L-methionine | | 分子名称: | NS5 methyltransferase, S-ADENOSYLMETHIONINE | | 著者 | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-01-16 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.507 Å) | | 主引用文献 | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
8JO0
 
 | |
2FP3
 
 | |
2G4A
 
 | |
8JNS
 
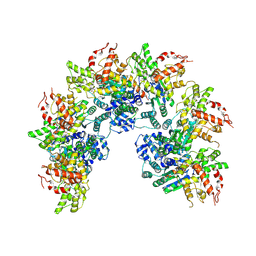 | | cryo-EM structure of a CED-4 hexamer | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | 著者 | Li, Y, Shi, Y. | | 登録日 | 2023-06-06 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural insights into CED-3 activation.
Life Sci Alliance, 6, 2023
|
|
4GJR
 
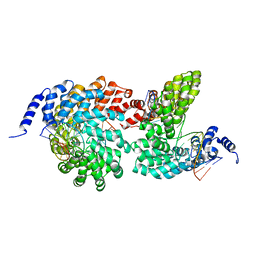 | | Crystal structure of the TAL effector dHax3 bound to methylated dsDNA | | 分子名称: | DNA (5'-D(*AP*GP*GP*GP*AP*GP*GP*TP*AP*GP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*(5CM)P*TP*AP*(5CM)P*CP*TP*CP*(5CM)P*CP*T)-3'), Hax3 | | 著者 | Yan, N, Deng, D, Yan, C.Y, Yin, P, Pan, X.J, Shi, Y.G. | | 登録日 | 2012-08-10 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of a protein complex
To be Published
|
|
6O0V
 
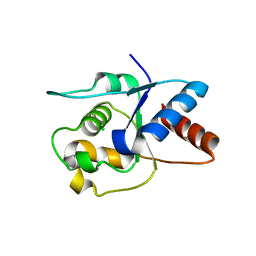 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 2 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1 | | 著者 | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | 登録日 | 2019-02-17 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0S
 
 | | Crystal structure of the tandem SAM domains from human SARM1 | | 分子名称: | Sterile alpha and TIR motif-containing protein 1 | | 著者 | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | 登録日 | 2019-02-17 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
7XQ8
 
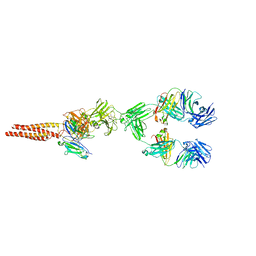 | | Structure of human B-cell antigen receptor of the IgM isotype | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, ... | | 著者 | Chen, M.Y, Su, Q, Shi, Y.G. | | 登録日 | 2022-05-07 | | 公開日 | 2022-08-17 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structure of the human IgM B cell receptor.
Science, 377, 2022
|
|
6TJX
 
 | | Cryo-EM structure of TypeII tau filaments extracted from the brains of individuals with Corticobasal degeneration | | 分子名称: | Microtubule-associated protein tau | | 著者 | Zhang, W, Murzin, A.G, Falcon, B, Shi, Y, Goedert, M, Scheres, S.H.W. | | 登録日 | 2019-11-27 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Novel tau filament fold in corticobasal degeneration.
Nature, 580, 2020
|
|
6O0T
 
 | | Crystal structure of selenomethionine labelled tandem SAM domains (L446M:L505M:L523M mutant) from human SARM1 | | 分子名称: | Sterile alpha and TIR motif-containing protein 1 | | 著者 | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | 登録日 | 2019-02-17 | | 公開日 | 2019-09-04 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O1B
 
 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 1 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sterile alpha and TIR motif-containing protein 1 | | 著者 | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | 登録日 | 2019-02-18 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0R
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with glycerol | | 分子名称: | GLYCEROL, Sterile alpha and TIR motif-containing protein 1 | | 著者 | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | 登録日 | 2019-02-17 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0Q
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with ribose | | 分子名称: | CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1, beta-D-ribofuranose | | 著者 | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | 登録日 | 2019-02-17 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
