4K2O
 
 | |
4K2P
 
 | |
7M03
 
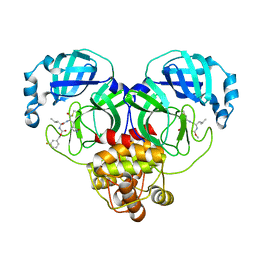 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 18c | | Descriptor: | (1R,2S)-2-((S)-2-((((3-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((3-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
2EXG
 
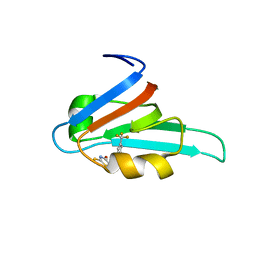 | | Making Protein-Protein Interactions Drugable: Discovery of Low-Molecular-Weight Ligands for the AF6 PDZ Domain | | Descriptor: | (5R)-2-SULFANYL-5-[4-(TRIFLUOROMETHYL)BENZYL]-1,3-THIAZOL-4-ONE, Afadin | | Authors: | Joshi, M, Vargas, C, Boisguerin, P, Krause, G, Schade, M, Oschkinat, H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Discovery of low-molecular-weight ligands for the AF6 PDZ domain.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
5FS4
 
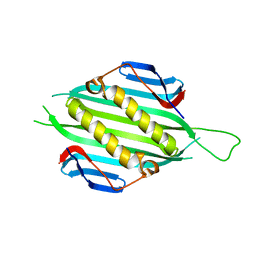 | | Bacteriophage AP205 coat protein | | Descriptor: | AP205 BACTERIOPHAGE COAT PROTEIN | | Authors: | Shishovs, M, Tars, K. | | Deposit date: | 2015-12-29 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of Ap205 Coat Protein Reveals Circular Permutation in Ssrna Bacteriophages.
J.Mol.Biol., 428, 2016
|
|
1MQ8
 
 | | Crystal structure of alphaL I domain in complex with ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, ... | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1LZ6
 
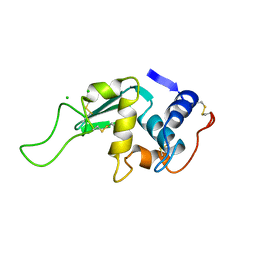 | | STRUCTURAL AND FUNCTIONAL ANALYSES OF THE ARG-GLY-ASP SEQUENCE INTRODUCED INTO HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, HUMAN LYSOZYME | | Authors: | Matsushima, M, Inaka, K, Yamada, T, Sekiguchi, K, Kikuchi, M. | | Deposit date: | 1993-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analyses of the Arg-Gly-Asp sequence introduced into human lysozyme.
J.Biol.Chem., 268, 1993
|
|
6J6Y
 
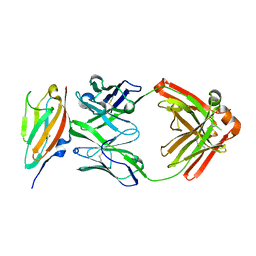 | | FGFR4 D2 - Fab complex | | Descriptor: | Fab Heavy chain, Fab light chain, Fibroblast growth factor receptor 4 | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Preclinical Development of U3-1784, a Novel FGFR4 Antibody Against Cancer, and Avoidance of Its On-target Toxicity.
Mol.Cancer Ther., 18, 2019
|
|
4G8W
 
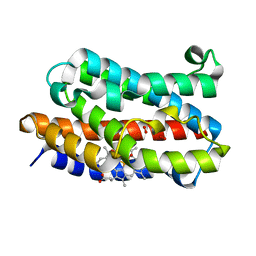 | | Rat Heme Oxygenase-1 in complex with Heme and O2 with 13 hr illumination: Laser on | | Descriptor: | FORMIC ACID, Heme oxygenase 1, OXYGEN MOLECULE, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
4G7T
 
 | | Rat Heme Oxygenase-1 in complex with Heme and CO with 1 hr Illumination: Laser on | | Descriptor: | CARBON MONOXIDE, FORMIC ACID, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-20 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
4G98
 
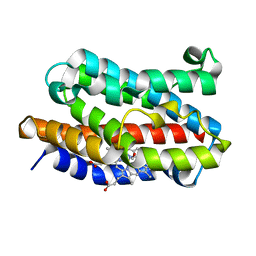 | | Rat Heme Oxygenase-1 in complex with Heme and CO at 100K | | Descriptor: | CARBON MONOXIDE, FORMIC ACID, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
4G8U
 
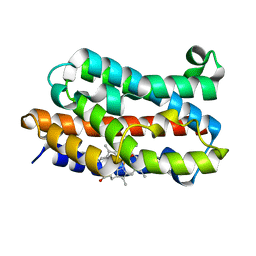 | | Rat Heme Oxygenase-1 in complex with Heme and O2 with 13 hr illumination: Laser off | | Descriptor: | FORMIC ACID, Heme oxygenase 1, OXYGEN MOLECULE, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
4G7P
 
 | | Rat Heme Oxygenase-1 in complex with Heme and CO with 1 hr Illumination at 100 K: Laser off | | Descriptor: | CARBON MONOXIDE, FORMIC ACID, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-20 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
4G8P
 
 | | Rat Heme Oxygenase-1 in complex with Heme and CO with 16 hr Illumination: Laser on | | Descriptor: | CARBON MONOXIDE, FORMIC ACID, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
2M7E
 
 | |
7LKS
 
 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2f | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKT
 
 | | 1.50 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2k | | Descriptor: | (1R,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
1C5H
 
 | | HYDROGEN BONDING AND CATALYSIS: AN UNEXPECTED EXPLANATION FOR HOW A SINGLE AMINO ACID SUBSTITUTION CAN CHANGE THE PH OPTIMUM OF A GLYCOSIDASE | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Joshi, M.D, Sidhu, G, Pot, I, Brayer, G.D, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 1999-11-24 | | Release date: | 2000-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase.
J.Mol.Biol., 299, 2000
|
|
4G7L
 
 | | Crystal Structure of rat Heme oxygenase-1 in complex with Heme and O2 | | Descriptor: | FORMIC ACID, Heme oxygenase 1, OXYGEN MOLECULE, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-20 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
1C5I
 
 | | HYDROGEN BONDING AND CATALYSIS: AN UNEXPECTED EXPLANATION FOR HOW A SINGLE AMINO ACID SUBSTITUTION CAN CHANGE THE PH OPTIMUM OF A GLYCOSIDASE | | Descriptor: | ENDO-1,4-BETA-XYLANASE, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Joshi, M.D, Sidhu, G, Pot, I, Brayer, G.D, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 1999-11-24 | | Release date: | 2000-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase.
J.Mol.Biol., 299, 2000
|
|
4G99
 
 | | Rat Heme Oxygenase-1 in complex with Heme and CO at 100 K after warming to 160 K | | Descriptor: | CARBON MONOXIDE, FORMIC ACID, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
2ZL9
 
 | | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation and crystal structure | | Descriptor: | (1R,2R,3R,5Z)-17-{(1S)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethoxy)-9,10-secoestra-5,7,16-triene-1,3-diol, Coactivator peptide DRIP, Vitamin D3 receptor | | Authors: | Shimizu, M, Miyamoto, Y, Nakabayashi, M, Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation, and crystal structure
Bioorg.Med.Chem., 16, 2008
|
|
2ZLC
 
 | | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation and crystal structure | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Coactivator peptide DRIP, Vitamin D3 receptor | | Authors: | Shimizu, M, Miyamoto, Y, Nakabayashi, M, Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation, and crystal structure
Bioorg.Med.Chem., 16, 2008
|
|
2ZLA
 
 | | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation and crystal structure | | Descriptor: | (1R,2S,3R,5Z,7E)-17-{(1R)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethoxy)-9,10-secoestra-5,7,16-triene-1,3-diol, Coactivator peptide DRIP, Vitamin D3 receptor | | Authors: | Shimizu, M, Miyamoto, Y, Nakabayashi, M, Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation, and crystal structure
Bioorg.Med.Chem., 16, 2008
|
|
7WOX
 
 | | PPARgamma antagonist (MMT-160)- PPARgamma LBD complex | | Descriptor: | N-[[5-(3-phenylprop-2-ynoylamino)-2-propoxy-phenyl]methyl]-4-pyrimidin-2-yl-benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Yoshizawa, M, Aoyama, T, Itoh, T, Miyachi, H. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Arylalkynyl amide-type peroxisome proliferator-activated receptor gamma (PPAR gamma )-selective antagonists covalently bind to the PPAR gamma ligand binding domain with a unique binding mode.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
