8ZJL
 
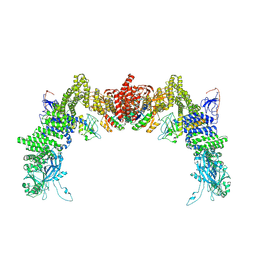 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 4) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJJ
 
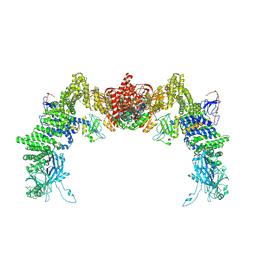 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 2) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJ2
 
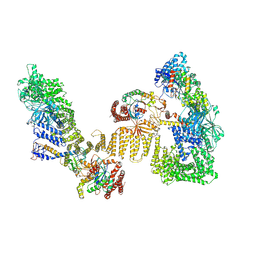 | | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJK
 
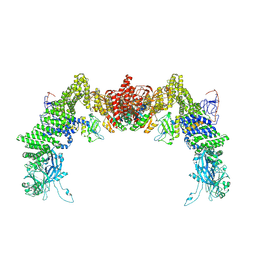 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 3) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJI
 
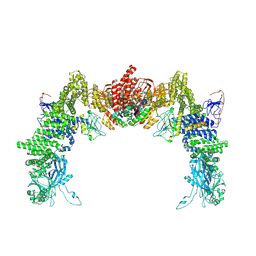 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 1) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJM
 
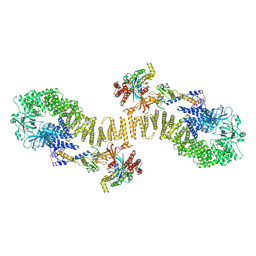 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 5) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
7K5I
 
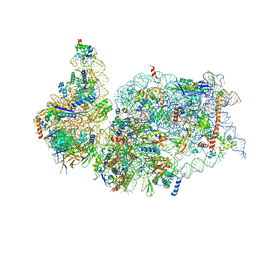 | | SARS-COV-2 nsp1 in complex with human 40S ribosome | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Wang, L, Shi, M, Wu, H. | | Deposit date: | 2020-09-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism.
Biorxiv, 2020
|
|
8XZ2
 
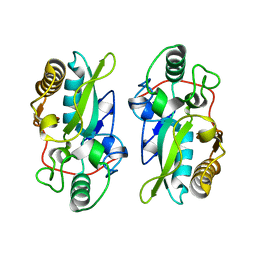 | | The structural model of a homodimeric D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria | | Descriptor: | D-alanyl-D-alanine dipeptidase | | Authors: | Konuma, T, Takai, T, Tsuchiya, C, Nishida, M, Hashiba, M, Yamada, Y, Shirai, H, Motoda, Y, Nagadoi, A, Chikaishi, E, Akagi, K, Akashi, S, Yamazaki, T, Akutsu, H, Oe, A, Ikegami, T. | | Deposit date: | 2024-01-20 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the homodimeric structure of a D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria.
Protein Sci., 33, 2024
|
|
3KDO
 
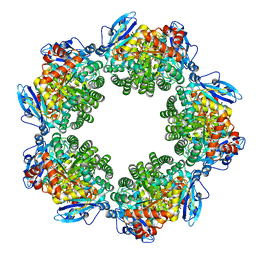 | | Crystal structure of Type III Rubisco SP6 mutant complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
3KDN
 
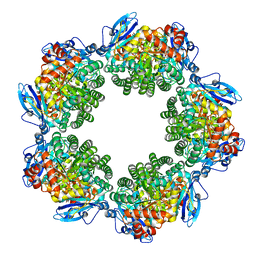 | | Crystal structure of Type III Rubisco SP4 mutant complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
1HJP
 
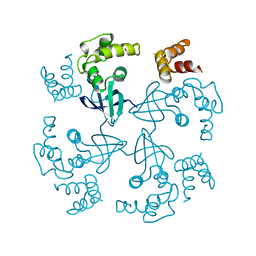 | | HOLLIDAY JUNCTION BINDING PROTEIN RUVA FROM E. COLI | | Descriptor: | RUVA | | Authors: | Nishino, T, Ariyoshi, M, Iwasaki, H, Shinagawa, H, Morikawa, K. | | Deposit date: | 1997-08-21 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Analyses of the Domain Structure in the Holliday Junction Binding Protein Ruva
Structure, 6, 1998
|
|
4V60
 
 | | The structure of rat liver vault at 3.5 angstrom resolution | | Descriptor: | Major vault protein | | Authors: | Kato, K, Zhou, Y, Tanaka, H, Yao, M, Yamashita, E, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2008-10-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of rat liver vault at 3.5 angstrom resolution
Science, 323, 2009
|
|
8IP8
 
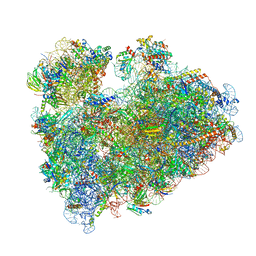 | | Wheat 80S ribosome stalled on AUG-Stop boron dependently | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eL8, 40S ribosomal protein eS1, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
2PSM
 
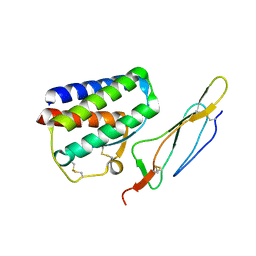 | | Crystal structure of Interleukin 15 in complex with Interleukin 15 receptor alpha | | Descriptor: | BENZAMIDINE, Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Olsen, S.K, Murayama, K, Kishishita, S, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Ota, N, Kanagawa, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Interleukin-15{middle dot}Interleukin-15 Receptor {alpha} Complex: INSIGHTS INTO TRANS AND CIS PRESENTATION
J.Biol.Chem., 282, 2007
|
|
6LPK
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 293 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
8J47
 
 | | CryoEM Structure of 40-Residue Arctic (E22G) Beta-Amyloid Fibril Derived by Co-Analysis with Solid-State NMR | E22G Abeta40 | | Descriptor: | E22G Amyloid-beta | | Authors: | Tehrani, M.J, Matsuda, I, Yamagata, A, Matsunaga, T, Sato, M, Toyooka, K, Shirouzu, M, Ishii, Y, Kodama, Y, McElheny, D, Kobayashi, N. | | Deposit date: | 2023-04-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | E22G A beta 40 fibril structure and kinetics illuminate how A beta 40 rather than A beta 42 triggers familial Alzheimer's.
Nat Commun, 15, 2024
|
|
5WVO
 
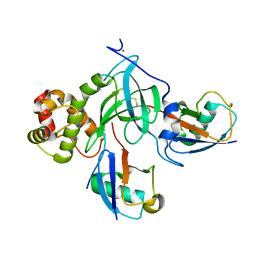 | | Crystal structure of DNMT1 RFTS domain in complex with K18/K23 mono-ubiquitylated histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Histone H3.1, Ubiquitin, ... | | Authors: | Ishiyama, S, Nishiyama, A, Nakanishi, M, Arita, K. | | Deposit date: | 2016-12-28 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance
Mol. Cell, 68, 2017
|
|
6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LPL
 
 | | A2AR crystallized in EROCOC17+4, SS-ROX at 100 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-11 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
7X73
 
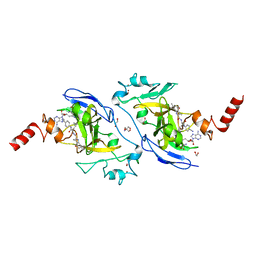 | | Structure of G9a in complex with RK-701 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A specific G9a inhibitor unveils BGLT3 lncRNA as a universal mediator of chemically induced fetal globin gene expression.
Nat Commun, 14, 2023
|
|
3JSV
 
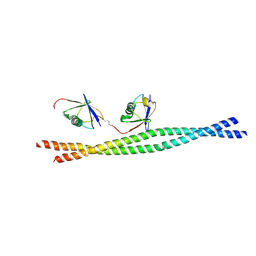 | | Crystal structure of mouse NEMO CoZi in complex with Lys63-linked di-ubiquitin | | Descriptor: | NF-kappa-B essential modulator, Ubiquitin | | Authors: | Yoshikawa, A, Sato, Y, Mimura, H, Yamashita, M, Yamagata, A, Fukai, S. | | Deposit date: | 2009-09-11 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin
Febs Lett., 583, 2009
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
7XN1
 
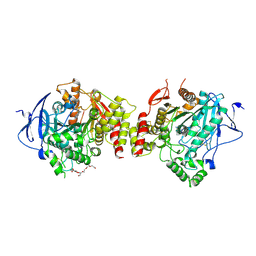 | | Crystal structure of human acetylcholinesterase in complex with tacrine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Acetylcholinesterase, ... | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2022-04-27 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
7XK5
 
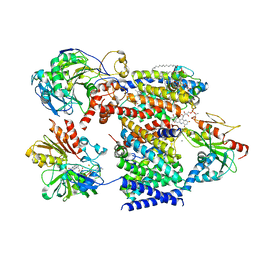 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 3 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
7XK6
 
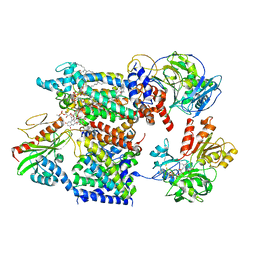 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, with aurachin D-42 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Aurachin D, CALCIUM ION, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
