2RUH
 
 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | Descriptor: | E3 ubiquitin-protein ligase Mdm2 | | Authors: | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
4U9H
 
 | | Ultra High Resolution Structure Of The Ni-R State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Nishikawa, K, Lubitz, W. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase.
Nature, 520, 2015
|
|
4TVT
 
 | | New ligand for thaumatin discovered using acoustic high throughput screening | | Descriptor: | ASCORBIC ACID, L(+)-TARTARIC ACID, SODIUM ION, ... | | Authors: | Teplitsky, E, Joshi, K, Ericson, D.L, Scalia, A, Mullen, J.D, Sweet, R.M, Soares, A.S. | | Deposit date: | 2014-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density.
J.Struct.Biol., 191, 2015
|
|
4U9I
 
 | | High Resolution Structure Of The Ni-R State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Nishikawa, K, Lubitz, W. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase.
Nature, 520, 2015
|
|
5B36
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with Cysteine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
7OWG
 
 | | human DEPTOR in a complex with mutant human mTORC1 A1459P | | Descriptor: | DEP domain-containing mTOR-interacting protein, Regulatory-associated protein of mTOR, Serine/threonine-protein kinase mTOR, ... | | Authors: | Heimhalt, M, Berndt, A, Wagstaff, J, Anandapadamanaban, M, Perisic, O, Maslen, S, McLaughlin, S, Yu, W.-H, Masson, G.R, Boland, A, Ni, X, Yamashita, K, Murshudov, G.N, Skehel, M, Freund, S.M, Williams, R.L. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Bipartite binding and partial inhibition links DEPTOR and mTOR in a mutually antagonistic embrace.
Elife, 10, 2021
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | Descriptor: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | Authors: | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
5B3A
 
 | | Crystal Structure of O-Phoshoserine Sulfhydrylase from Aeropyrum pernix in Complexed with the alpha-Aminoacrylate Intermediate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Protein CysO | | Authors: | Nakamura, T, Takeda, E, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2016-02-12 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Role of F225 in O-phosphoserine sulfhydrylase from Aeropyrum pernix K1
Extremophiles, 20, 2016
|
|
5MR2
 
 | | Crystal structure of red abalone VERL repeat 2 with linker at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Vitelline envelope sperm lysin receptor | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5MP7
 
 | | Crystal structure of phosphoribosylpyrophosphate synthetase from Mycobacterium smegmatis | | Descriptor: | ACETATE ION, Ribose-phosphate pyrophosphokinase | | Authors: | Donini, S, Garavaglia, S, Ferraris, D.M, Miggiano, R, Mori, S, Shibayama, K, Rizzi, M. | | Deposit date: | 2016-12-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural investigations on phosphoribosylpyrophosphate synthetase from Mycobacterium smegmatis.
PLoS ONE, 12, 2017
|
|
1IPA
 
 | | CRYSTAL STRUCTURE OF RNA 2'-O RIBOSE METHYLTRANSFERASE | | Descriptor: | RNA 2'-O-RIBOSE METHYLTRANSFERASE | | Authors: | Nureki, O, Shirouzu, M, Hashimoto, K, Ishitani, R, Terada, T, Tamakoshi, M, Oshima, T, Chijimatsu, M, Takio, K, Vassylyev, D.G, Shibata, T, Inoue, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An enzyme with a deep trefoil knot for the active-site architecture.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1A99
 
 | | PUTRESCINE RECEPTOR (POTF) FROM E. COLI | | Descriptor: | 1,4-DIAMINOBUTANE, PUTRESCINE-BINDING PROTEIN | | Authors: | Vassylyev, D.G, Tomitori, H, Kashiwagi, K, Morikawa, K, Igarashi, K. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and mutational analysis of the Escherichia coli putrescine receptor. Structural basis for substrate specificity.
J.Biol.Chem., 273, 1998
|
|
6OON
 
 | | Human Argonaute4 bound to guide RNA | | Descriptor: | Protein argonaute-4, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3') | | Authors: | Park, M.S, Brackbill, J.A, Nakanishi, K. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multidomain Convergence of Argonaute during RISC Assembly Correlates with the Formation of Internal Water Clusters.
Mol.Cell, 75, 2019
|
|
7V94
 
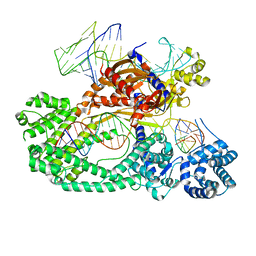 | | Cryo-EM structure of the Cas12c2-sgRNA-target DNA ternary complex | | Descriptor: | Cas12c2, sgRNA, target DNA (non target strand), ... | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
7V93
 
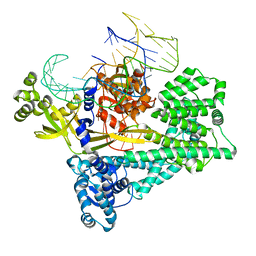 | | Cryo-EM structure of the Cas12c2-sgRNA binary complex | | Descriptor: | cas12c2, sgRNA | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
1NBK
 
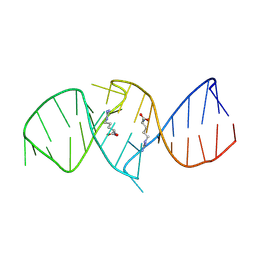 | | The structure of RNA aptamer for HIV Tat complexed with two argininamide molecules | | Descriptor: | 2-AMINO-5-GUANIDINO-PENTANOIC ACID, RNA aptamer | | Authors: | Matsugami, A, Kobayashi, S, Ouhashi, K, Uesugi, S, Yamamoto, R, Taira, K, Nishikawa, S, Kumar, P.K.R, Katahira, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2024-09-18 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Highly Efficient Trapping of the HIV Tat Protein by an RNA Aptamer
Structure, 11, 2003
|
|
8JHH
 
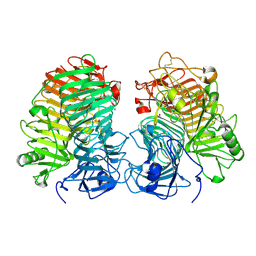 | | Glycoside hydrolase family 55 endo-beta-1,3-glucanase from Microdochium nivale | | Descriptor: | GLYCEROL, MnLam55A | | Authors: | Ota, T, Saburi, W, Yamashita, K, Tagami, T, Yu, J, Komba, S, Jewell, L.E, Hsiang, T, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2023-05-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for endo-type action of glycoside hydrolase family 55 endo-beta-1,3-glucanase on beta 1-3/1-6-glucan.
J.Biol.Chem., 299, 2023
|
|
1F54
 
 | | SOLUTION STRUCTURE OF THE APO N-TERMINAL DOMAIN OF YEAST CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2000-06-13 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
1F55
 
 | | SOLUTION STRUCTURE OF THE CALCIUM BOUND N-TERMINAL DOMAIN OF YEAST CALMODULIN | | Descriptor: | CALCIUM ION, CALMODULIN | | Authors: | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2000-06-13 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
6O5E
 
 | | Crystal structure of the Vitronectin hemopexin-like domain | | Descriptor: | CHLORIDE ION, IMIDAZOLE, NITRATE ION, ... | | Authors: | Lechtenberg, B.C, Shin, K, Marassi, F.M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human Vitronectin C-terminal domain and interaction withYersinia pestisouter membrane protein Ail.
Sci Adv, 5, 2019
|
|
3MBE
 
 | | TCR 21.30 in complex with MHC class II I-Ag7HEL(11-27) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II H2-IAg7 ALPHA CHAIN, ... | | Authors: | Corper, A.L, Yoshida, K, Teyton, L, Wilson, I.A. | | Deposit date: | 2010-03-25 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | The diabetogenic mouse MHC class II molecule I-Ag7 is endowed with a switch that modulates TCR affinity.
J.Clin.Invest., 120, 2010
|
|
3O15
 
 | | Crystal Structure of Bacillus subtilis Thiamin Phosphate Synthase Complexed with a Carboxylated Thiazole Phosphate | | Descriptor: | 2-TRIFLUOROMETHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-methyl-5-[2-(phosphonooxy)ethyl]-1,3-thiazole-2-carboxylic acid, PYROPHOSPHATE 2-, ... | | Authors: | McCulloch, K.M, Hanes, J.W, Abdelwahed, S, Mahanta, N, Hazra, A, Ishida, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-07-20 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Kinetic Characterization of Bacillus subtilis
Thiamin Phosphate Synthase with a Carboxylated Thiazole Phosphate
to be published
|
|
