7XHR
 
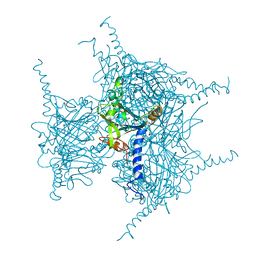 | | Crystal structure of Wild Type Cypovirus Polyhedra produced by cell-free protein synthesis | | Descriptor: | ACETYL GROUP, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tanaka, J, Kojima, M, Hirata, K, Yamashita, K, Ueno, T. | | Deposit date: | 2022-04-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Cell-free protein crystallization for nanocrystal structure determination.
Sci Rep, 12, 2022
|
|
7WYR
 
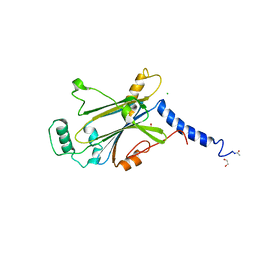 | | Crystal structure of Cypovirus Polyhedra mutant fused with CLN025 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin fused with CLN025 | | Authors: | Kojima, M, Abe, S, Hirata, K, Yamashita, K, Ueno, T. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering of an in-cell protein crystal for fastening a metastable conformation of a target miniprotein.
Biomater Sci, 11, 2023
|
|
5YC8
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS (Hg-derivative) | | Descriptor: | MERCURY (II) ION, Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
7YF6
 
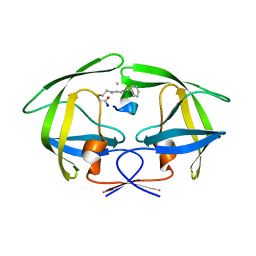 | | Crystal structure of HIV-1 protease in complex with macrocyclic peptide | | Descriptor: | MAGNESIUM ION, Macrocyclic Peptide, Protease | | Authors: | Kusumoto, Y, Sato, S, Yamada, T, Kozono, I, Nakata, Z, Asada, N, Mitsuki, S, Wakasa-Morimoto, C, Tohru, M, Watanabe, A, Hayashi, K, Mikamiyama, H. | | Deposit date: | 2022-07-07 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Highly Potent and Oral Macrocyclic Peptides as a HIV-1 Protease Inhibitor: mRNA Display-Derived Hit-to-Lead Optimization.
Acs Med.Chem.Lett., 13, 2022
|
|
5ZK8
 
 | | Crystal structure of M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
3EQN
 
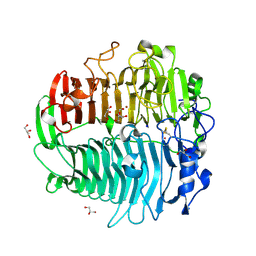 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) | | Descriptor: | ACETATE ION, GLYCEROL, Glucan 1,3-beta-glucosidase, ... | | Authors: | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
5ZIH
 
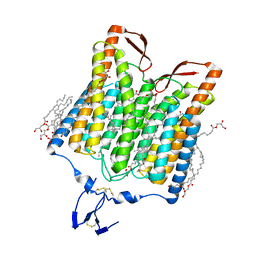 | | Crystal structure of the red light-activated channelrhodopsin Chrimson. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sensory opsin A,Chrimson | | Authors: | Oda, K, Vierock, J, Oishi, S, Taniguchi, R, Yamashita, K, Nishizawa, T, Hegemann, P, Nureki, O. | | Deposit date: | 2018-03-15 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the red light-activated channelrhodopsin Chrimson.
Nat Commun, 9, 2018
|
|
5ZKB
 
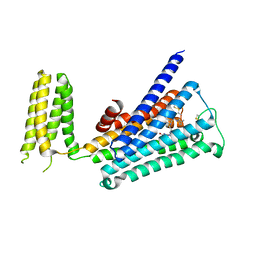 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with AF-DX 384 | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-[2-[(2S)-2-[(dipropylamino)methyl]piperidin-1-yl]ethyl]-6-oxidanylidene-5H-pyrido[2,3-b][1,4]benzodiazepine-11-carboxamide | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZKC
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZK3
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with QNB | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2 | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5AZ2
 
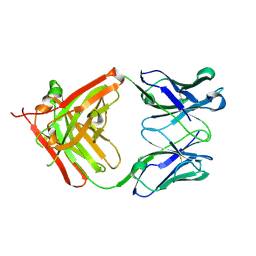 | | Crystal structure of the Fab fragment of 9E5, a murine monoclonal antibody specific for human epiregulin | | Descriptor: | anti-human epiregulin antibody 9E5 Fab heavy chain, anti-human epiregulin antibody 9E5 Fab light chain | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
7YML
 
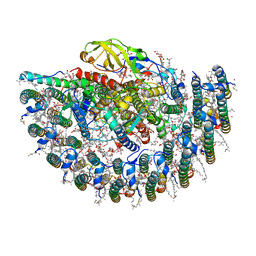 | | Structure of photosynthetic LH1-RC super-complex of Rhodobacter capsulatus | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tani, K, Kanno, R, Ji, X.-C, Satoh, I, Kobayashi, Y, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Mizoguchi, A, Humbel, B.M, Madigan, M.T, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Rhodobacter capsulatus forms a compact crescent-shaped LH1-RC photocomplex.
Nat Commun, 14, 2023
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | Descriptor: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | Authors: | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
5B37
 
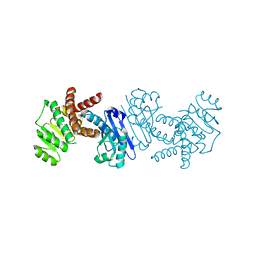 | | Crystal structure of L-tryptophan dehydrogenase from Nostoc punctiforme | | Descriptor: | Tryptophan dehydrogenase | | Authors: | Wakamatsu, T, Sakuraba, H, Kitamura, M, Hakumai, Y, Ohnishi, K, Ashiuchi, M, Ohshima, T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Insights into l-Tryptophan Dehydrogenase from a Photoautotrophic Cyanobacterium, Nostoc punctiforme.
Appl. Environ. Microbiol., 83, 2017
|
|
3EQO
 
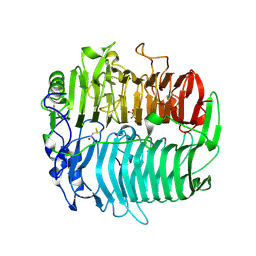 | | Crystal structure of beta-1,3-glucanase from Phanerochaete chrysosporium (Lam55A) gluconolactone complex | | Descriptor: | D-glucono-1,5-lactone, Glucan 1,3-beta-glucosidase, ZINC ION, ... | | Authors: | Ishida, T, Fushinobu, S, Kawai, R, Kitaoka, M, Igarashi, K, Samejima, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of glycoside hydrolase family 55 beta -1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
J.Biol.Chem., 284, 2009
|
|
8JHH
 
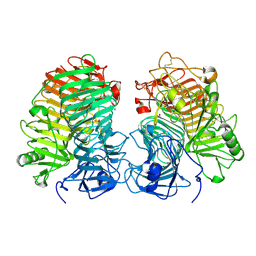 | | Glycoside hydrolase family 55 endo-beta-1,3-glucanase from Microdochium nivale | | Descriptor: | GLYCEROL, MnLam55A | | Authors: | Ota, T, Saburi, W, Yamashita, K, Tagami, T, Yu, J, Komba, S, Jewell, L.E, Hsiang, T, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2023-05-23 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for endo-type action of glycoside hydrolase family 55 endo-beta-1,3-glucanase on beta 1-3/1-6-glucan.
J.Biol.Chem., 299, 2023
|
|
5H0Q
 
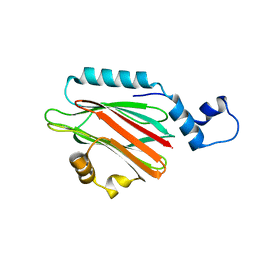 | | Crystal structure of lipid binding protein Nakanori at 1.5A | | Descriptor: | Lipid binding protein | | Authors: | Makino, A, Abe, M, Ishitsuka, R, Murate, M, Kishimoto, T, Sakai, S, Hullin-Matsuda, F, Shimada, Y, Inaba, T, Miyatake, H, Tanaka, H, Kurahashi, A, Pack, C.G, Kasai, R.S, Kubo, S, Schieber, N.L, Dohmae, N, Tochio, N, Hagiwara, K, Sasaki, Y, Aida, Y, Fujimori, F, Kigawa, T, Nishikori, K, Parton, R.G, Kusumi, A, Sako, Y, Anderluh, G, Yamashita, M, Kobayashi, T, Greimel, P, Kobayashi, T. | | Deposit date: | 2016-10-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C
FASEB J., 31, 2017
|
|
5B2C
 
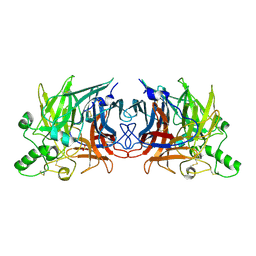 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, SULFATE ION | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B2D
 
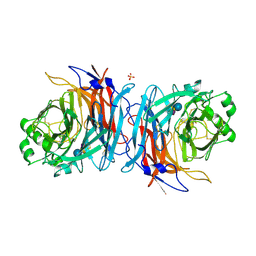 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to 3-sialyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6LEO
 
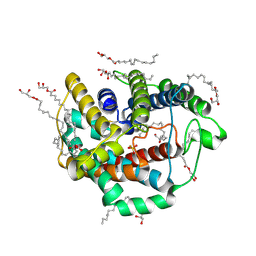 | | Crystal structure of thiosulfate transporter YeeE from Spirochaeta thermophila | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Takeuchi, A, Uchino, S, Sugano, Y. | | Deposit date: | 2019-11-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
5OMD
 
 | | Crystal structure of S. cerevisiae Ddc2 N-terminal coiled-coil domain | | Descriptor: | DNA damage checkpoint protein LCD1 | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
5WY9
 
 | |
5CPG
 
 | | R-Hydratase PhaJ1 from Pseudomonas aeruginosa in the unliganded form | | Descriptor: | (R)-specific enoyl-CoA hydratase, GLYCEROL | | Authors: | Tsuge, T, Sato, S, Hiroe, A, Ishizuka, K, Kanazawa, H, Kanagarajan, S, Shiro, Y, Hisano, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Contribution of the Distal Pocket Residue to the Acyl-Chain-Length Specificity of (R)-Specific Enoyl-Coenzyme A Hydratases from Pseudomonas spp.
Appl.Environ.Microbiol., 81, 2015
|
|
6LEP
 
 | | Crystal structure of thiosulfate transporter YeeE inactive mutant - C91A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Sugano, Y, Takeuchi, A, Uchino, S. | | Deposit date: | 2019-11-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
6LUB
 
 | | Crystal Structure of EGFR(L858R/T790M/C797S) in complex with CH7233163 | | Descriptor: | Epidermal growth factor receptor, N-[2-(1-cyclopropylsulfonylpyrazol-4-yl)pyrimidin-4-yl]-7-(4-methylpiperazin-1-yl)-5-propan-2-yl-9-[2,2,2-tris(fluoranyl)ethoxy]pyrido[4,3-b]indol-3-amine | | Authors: | Kawauchi, H, Fukami, T.A, Sato, S, Endo, M, Torizawa, T, Kashima, K, Chiba, T, Sakamoto, H. | | Deposit date: | 2020-01-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | CH7233163 Overcomes Osimertinib-Resistant EGFR-Del19/T790M/C797S Mutation.
Mol.Cancer Ther., 19, 2020
|
|
