5FHI
 
 | | Crystallographic structure of PsoE without Co | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, putative | | Authors: | Hara, K, Hashimoto, H, Yamamoto, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2015-12-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Oxidative trans to cis Isomerization of Olefins in Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
4WFJ
 
 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 1.75 angstrom resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cutinase | | Authors: | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WFK
 
 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 2.35 angstrom resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cutinase | | Authors: | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WFI
 
 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-free state | | Descriptor: | Cutinase | | Authors: | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
2RUH
 
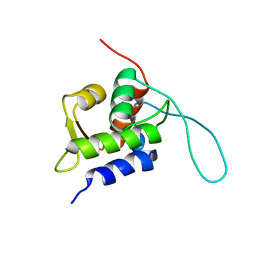 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | Descriptor: | E3 ubiquitin-protein ligase Mdm2 | | Authors: | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
8WUC
 
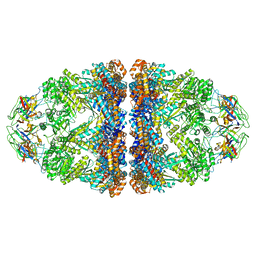 | | Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
4XVD
 
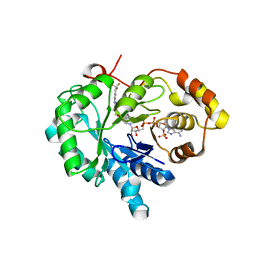 | | 17beta-HSD5 in complex with 4-nitro-2-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}methyl)phenol | | Descriptor: | 4-nitro-2-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}methyl)phenol, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Amano, Y, Yamaguchi, T, Niimi, T, Sakashita, H. | | Deposit date: | 2015-01-27 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8FJN
 
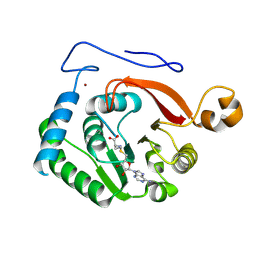 | | Crystal Structure of the Trypanosoma brucei DOT1A histone H3K76 methyltransferase in complex with AdoHcy - C2221 space group | | Descriptor: | CALCIUM ION, CHLORIDE ION, Histone-lysine N-methyltransferase, ... | | Authors: | Frisbie, V.S, Hashimoto, H, Debler, E.W. | | Deposit date: | 2022-12-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two DOT1 enzymes cooperatively mediate efficient ubiquitin-independent histone H3 lysine 76 tri-methylation in kinetoplastids.
Nat Commun, 15, 2024
|
|
8FJM
 
 | | Crystal Structure of the Trypanosoma brucei DOT1A histone H3K76 methyltransferase in complex with AdoHcy - P212121 space group | | Descriptor: | ACETATE ION, CALCIUM ION, Histone-lysine N-methyltransferase, ... | | Authors: | Frisbie, V.S, Hashimoto, H, Debler, E.W. | | Deposit date: | 2022-12-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two DOT1 enzymes cooperatively mediate efficient ubiquitin-independent histone H3 lysine 76 tri-methylation in kinetoplastids.
Nat Commun, 15, 2024
|
|
8GST
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (pyruvate bound-form) | | Descriptor: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION, PYRUVIC ACID | | Authors: | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
8GSR
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (apo-form) | | Descriptor: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION | | Authors: | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
8GNN
 
 | | Crystal structure of the human RAD9-RAD1-HUS1-RAD17 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Cell cycle checkpoint protein RAD17, ... | | Authors: | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | The 9-1-1 DNA clamp subunit RAD1 forms specific interactions with clamp loader RAD17, revealing functional implications for binding-protein RHINO.
J.Biol.Chem., 299, 2023
|
|
6M30
 
 | | Crystal structure of a mutant Staphylococcus equorum manganese superoxide dismutase N73F | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Retnoningrum, D.S, Yoshida, H, Razani, M.D, Meidianto, V.F, Hartanto, A, Artarini, A, Ismaya, W.T. | | Deposit date: | 2020-03-02 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Unprecedented Role of The N73-F124 Pair in The Staphylococcus equorum MnSOD Activity.
Curr Enzym Inhib, 2021
|
|
6PH2
 
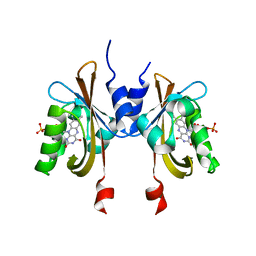 | | Complete LOV domain from the LOV-HK sensory protein from Brucella abortus (mutant C69S, construct 15-155) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PH4
 
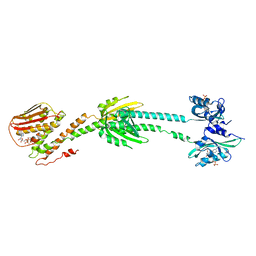 | | Full length LOV-PAS-HK construct from the LOV-HK sensory protein from Brucella abortus (light-adapted, construct 15-489) | | Descriptor: | Blue-light-activated histidine kinase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PH3
 
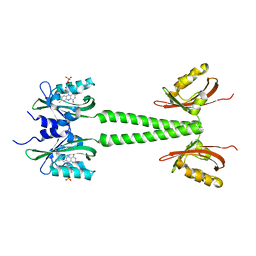 | | LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (dark-adapted, construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PPS
 
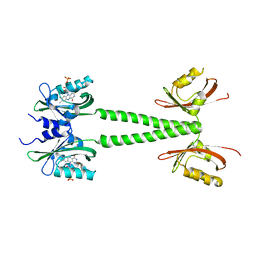 | | A blue light illuminated LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Fernandez, I, Shin, H, Gunawardana, S, Otero, L.H, Cerutti, M.L, Yang, X, Klinke, S, Goldbaum, F.A. | | Deposit date: | 2019-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
7E6Z
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 50 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6Y
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6X
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 4 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E71
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E70
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 250 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
8RXR
 
 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | Descriptor: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 18, 2024
|
|
5B2T
 
 | | Crystal structure of the Streptococcus pyogenes Cas9 VRER variant in complex with sgRNA and target DNA (TGCG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
1MG8
 
 | | NMR structure of ubiquitin-like domain in murine Parkin | | Descriptor: | Parkin | | Authors: | Tashiro, M, Okubo, S, Shimotakahara, S, Hatanaka, H, Yasuda, H, Kainosho, M, Yokoyama, S, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-15 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of ubiquitin-like domain in PARKIN: Gene product of familial Parkinson's disease.
J.Biomol.NMR, 25, 2003
|
|
