6AKI
 
 | | Calcium release-activated calcium channel protein 1, P288L mutant | | Descriptor: | CHLORIDE ION, Calcium release-activated calcium channel protein 1 | | Authors: | Liu, X, Wu, G, Yang, X, Shen, Y. | | Deposit date: | 2018-09-01 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (4.496 Å) | | Cite: | Calcium release-activated calcium channel protein 1, P288L mutant
To Be Published
|
|
4L16
 
 | | Crystal structure of FIGL-1 AAA domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fidgetin-like protein 1 | | Authors: | Peng, W, Lin, Z, Li, W, Lu, J, Shen, Y, Wang, C. | | Deposit date: | 2013-06-02 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein.
J.Biol.Chem., 288, 2013
|
|
6JAU
 
 | | The complex structure of Pseudomonas aeruginosa MucA/MucB. | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Li, T, He, L.H, Li, C.C, Liu, L, Peng, C.T, Shen, Y.L, Qin, X.F, Xiao, Q.J, Zhu, Y.B, Song, Y.J, Zhao, N.l, Zhao, C, Yang, J, Mu, X.Y, Huang, Q, Bao, R. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Molecular basis of the lipid-induced MucA-MucB dissociation in Pseudomonas aeruginosa.
Commun Biol, 3, 2020
|
|
4L15
 
 | | Crystal structure of FIGL-1 AAA domain | | Descriptor: | Fidgetin-like protein 1, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Peng, W, Lin, Z, Li, W, Lu, J, Shen, Y, Wang, C. | | Deposit date: | 2013-06-02 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein.
J.Biol.Chem., 288, 2013
|
|
6BEK
 
 | |
3TEQ
 
 | | Crystal structure of SOAR domain | | Descriptor: | PHOSPHATE ION, Stromal interaction molecule 1 | | Authors: | Yang, X, Jin, H, Cai, X, Shen, Y. | | Deposit date: | 2011-08-15 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the activation of Stromal interaction molecule 1 (STIM1).
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3LRA
 
 | | Structural Basis for Assembling a Human Tripartite Complex Dlg1-MPP7-Mals3 | | Descriptor: | Disks large homolog 1, MAGUK p55 subfamily member 7, Protein lin-7 homolog C | | Authors: | Yang, X, Xie, X, Shen, Y, Long, J. | | Deposit date: | 2010-02-10 | | Release date: | 2010-11-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for tandem L27 domain-mediated polymerization
Faseb J., 24, 2010
|
|
2HGA
 
 | | Solution NMR Structure of Conserved protein MTH1368, Northeast Structural Genomics Consortium Target TT821A | | Descriptor: | Conserved protein MTH1368 | | Authors: | Liu, G, Lin, Y, Parish, D, Shen, Y, Sukumaran, D, Yee, A, Semesi, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Conserved protein MTH1368, Northeast Structural Genomics Consortium Target TT821A
TO BE PUBLISHED
|
|
5Z6R
 
 | | SPASTIN AAA WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Spastin | | Authors: | Lin, Z, Wang, C, Shen, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Aaa Family Protein Spastin Severs A Microtubule
To Be Published
|
|
3TER
 
 | |
5YWW
 
 | | Archael RuvB-like Holiday junction helicase | | Descriptor: | GLYCEROL, Nucleotide binding protein PINc | | Authors: | Zhai, B, Yuan, Z, Han, X, DuPrez, K, Shen, Y, Fan, L. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The archaeal ATPase PINA interacts with the helicase Hjm via its carboxyl terminal KH domain remodeling and processing replication fork and Holliday junction.
Nucleic Acids Res., 46, 2018
|
|
3TUO
 
 | |
4HRV
 
 | |
4I16
 
 | | Crystal structure of CARMA1 CARD | | Descriptor: | Caspase recruitment domain-containing protein 11, SULFATE ION | | Authors: | Li, S, Yang, X, Shen, Y. | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural insights into the assembly of CARMA1 and BCL10
Plos One, 7, 2012
|
|
2JBU
 
 | | Crystal structure of human insulin degrading enzyme complexed with co- purified peptides. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CO-PURIFIED PEPTIDE, INSULIN-DEGRADING ENZYME | | Authors: | Im, H, Shen, Y, Tang, W.J. | | Deposit date: | 2006-12-11 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
6KNM
 
 | | Apelin receptor in complex with single domain antibody | | Descriptor: | Apelin receptor,Rubredoxin,Apelin receptor, Single domain antibody JN241, ZINC ION | | Authors: | Ma, Y.B, Ding, Y, Song, X, Ma, X, Li, X, Zhang, N, Song, Y, Sun, Y, Shen, Y, Zhong, W, Hu, L.A, Ma, Y.L, Zhang, M.Y. | | Deposit date: | 2019-08-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-guided discovery of a single-domain antibody agonist against human apelin receptor.
Sci Adv, 6, 2020
|
|
6IGB
 
 | | the structure of Pseudomonas aeruginosa Periplasmic gluconolactonase, PpgL | | Descriptor: | ACETATE ION, Periplasmic gluconolactonase, PpgL, ... | | Authors: | Song, Y.J, Shen, Y.L, Wang, K.L, Li, T, Zhu, Y.B, Li, C.C, He, L.H, Zhao, N.L, Zhao, C, Yang, J, Huang, Q, Mu, X.Y. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and Functional Insights into PpgL, a Metal-Independent beta-Propeller Gluconolactonase That Contributes toPseudomonas aeruginosaVirulence.
Infect.Immun., 87, 2019
|
|
2IL1
 
 | | Crystal structure of a predicted human GTPase in complex with GDP | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, J, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a predicted human GTPase in complex with GDP
To be Published
|
|
2G99
 
 | |
8IP4
 
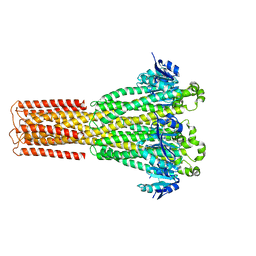 | | Cryo-EM structure of hMRS-highEDTA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP5
 
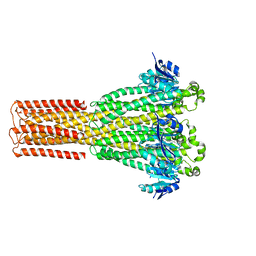 | | Cryo-EM structure of hMRS2-lowEDTA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP6
 
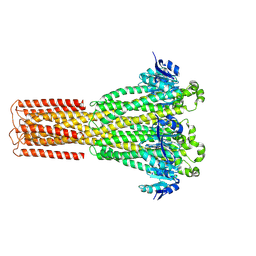 | | Cryo-EM structure of hMRS2-rest | | Descriptor: | CHLORIDE ION, Magnesium transporter MRS2 homolog, mitochondrial | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP3
 
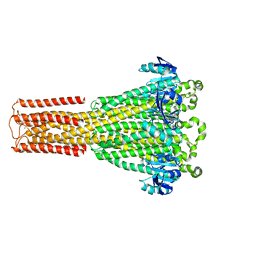 | | Cryo-EM structure of hMRS2-Mg | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
2G9A
 
 | |
2J9J
 
 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2006-11-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
