7MUC
 
 | | Legionella pneumophila Dot/Icm T4SS C1 Reconstruction | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUS
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 2 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
7MUE
 
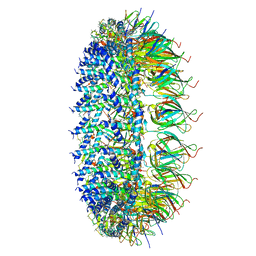 | | Legionella pneumophila Dot/Icm T4SS PR | | Descriptor: | DotF, IcmE protein, Type IV secretion protein IcmK, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
5BQI
 
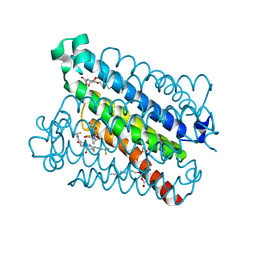 | | Discovery of a Potent and Selective mPGES-1 Inhibitor for the Treatment of Pain | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 2-(difluoromethyl)-5-{[(2-methylpropanoyl)amino]methyl}-N-{5-methyl-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-2-yl}pyridine-3-carboxamide, GLUTATHIONE, ... | | Authors: | Fisher, M.J, Schiffler, M.A, Kuklish, S.L, Antonysamy, S, Luz, J.G. | | Deposit date: | 2015-05-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors.
J.Med.Chem., 59, 2016
|
|
5BQH
 
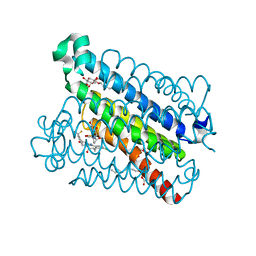 | | Discovery of a Potent and Selective mPGES-1 Inhibitor for the Treatment of Pain | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, GLUTATHIONE, N-[4-(4-chlorophenyl)-1H-imidazol-2-yl]-2-(difluoromethyl)-5-{[(2-methylpropanoyl)amino]methyl}benzamide, ... | | Authors: | Fisher, M.J, Schiffler, M.A, Kuklish, S.L, Antonysamy, S, Luz, J.G. | | Deposit date: | 2015-05-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors.
J.Med.Chem., 59, 2016
|
|
5BQG
 
 | | Crystal Structure of mPGES-1 Bound to an Inhibitor | | Descriptor: | 2-chloro-N-(4-phenyl-1,3-thiazol-2-yl)benzamide, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Fisher, M.J, Schiffler, M.A, Kuklish, S.L, Antonysamy, S, Luz, J.G. | | Deposit date: | 2015-05-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.436 Å) | | Cite: | Discovery and Characterization of 2-Acylaminoimidazole Microsomal Prostaglandin E Synthase-1 Inhibitors.
J.Med.Chem., 59, 2016
|
|
7EIM
 
 | |
7EJT
 
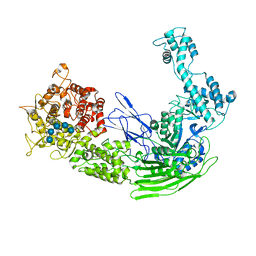 | |
7EJP
 
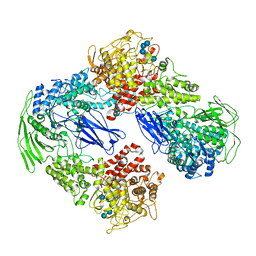 | |
7EKX
 
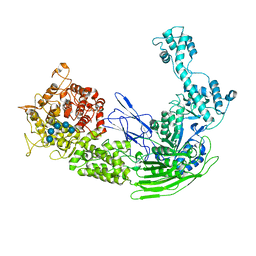 | |
7EKW
 
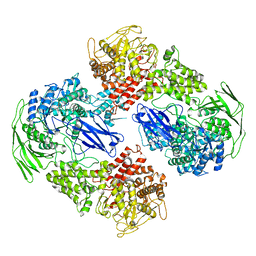 | |
7EKU
 
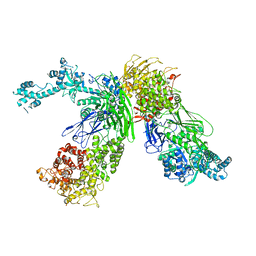 | |
4GXZ
 
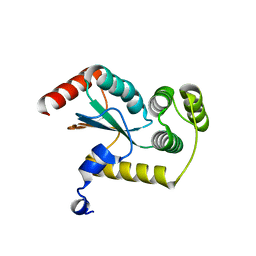 | | Crystal structure of a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium | | Descriptor: | Suppression of copper sensitivity protein | | Authors: | Shepherd, M, Heras, B, King, G.J, Argente, M.P, Achard, M.E.S, King, N.P, McEwan, A.G, Schembri, M.A. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and functional characterization of ScsC, a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium
Antioxid Redox Signal, 19, 2013
|
|
5CRA
 
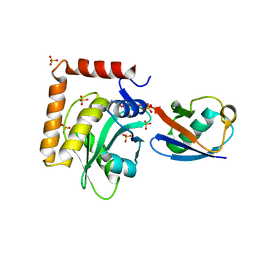 | | Structure of the SdeA DUB Domain | | Descriptor: | METHYL 4-AMINOBUTANOATE, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Sheedlo, M.J, Qiu, J, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2AYW
 
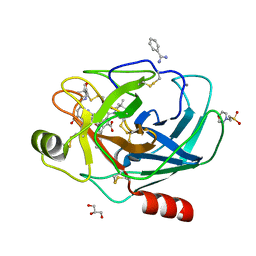 | | Crystal Structure of the complex formed between trypsin and a designed synthetic highly potent inhibitor in the presence of benzamidine at 0.97 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-({[4-(DIAMINOMETHYL)PHENYL]AMINO}CARBONYL)-6-METHOXYPYRIDIN-3-YL]-5-{[(1-FORMYL-2,2-DIMETHYLPROPYL)AMINO]CARBONYL}BENZOIC ACID, BENZAMIDINE, ... | | Authors: | Sherawat, M, Kaur, P, Perbandt, M, Betzel, C, Slusarchyk, W.A, Bisacchi, G.S, Chang, C, Jacobson, B.L, Einspahr, H.M, Singh, T.P. | | Deposit date: | 2005-09-09 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of the complex of trypsin with a highly potent synthetic inhibitor at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4LMO
 
 | |
6MRM
 
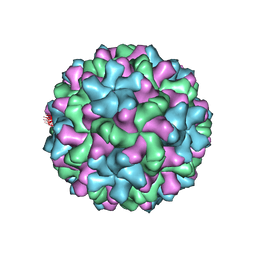 | | Red Clover Necrotic Mosaic Virus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Sherman, M.B, Smith, T.J. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Near-Atomic-Resolution Cryo-Electron Microscopy Structures of Cucumber Leaf Spot Virus and Red Clover Necrotic Mosaic Virus: Evolutionary Divergence at the Icosahedral Three-Fold Axes.
J.Virol., 94, 2020
|
|
6MRL
 
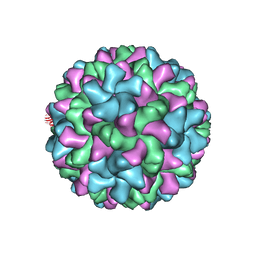 | | Cucumber Leaf Spot Virus | | Descriptor: | CALCIUM ION, p41 | | Authors: | Sherman, M.B, Smith, T.J. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Near-Atomic-Resolution Cryo-Electron Microscopy Structures of Cucumber Leaf Spot Virus and Red Clover Necrotic Mosaic Virus: Evolutionary Divergence at the Icosahedral Three-Fold Axes.
J.Virol., 94, 2020
|
|
4JCX
 
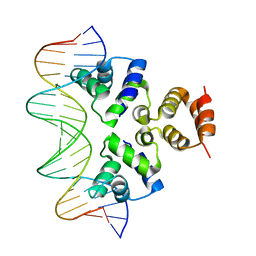 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OL operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*CP*AP*CP*TP*AP*AP*GP*GP*AP*AP*AP*AP*CP*TP*TP*AP*GP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*AP*AP*GP*TP*TP*TP*TP*CP*CP*TP*TP*AP*GP*TP*GP*T)-3') | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, Mcgeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5CRC
 
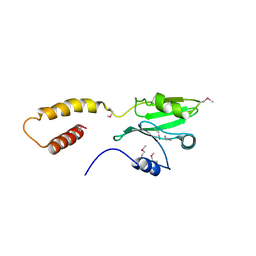 | | Structure of the SdeA DUB Domain | | Descriptor: | SdeA | | Authors: | Sheedlo, M.J, Qiu, J, Tan, Y, Paul, L.N, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CRB
 
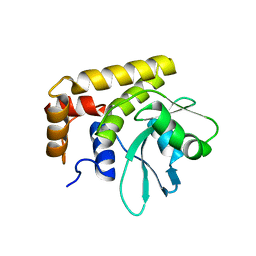 | | Crystal Structure of SdeA DUB | | Descriptor: | SdeA | | Authors: | Sheedlo, M.J, Qiu, J, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4JQD
 
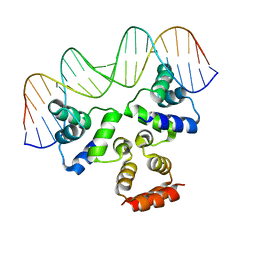 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OL operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*CP*AP*CP*TP*AP*AP*GP*GP*AP*AP*AP*AP*CP*TP*TP*AP*GP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*AP*AP*GP*TP*TP*TP*TP*CP*CP*TP*TP*AP*GP*TP*GP*T)-3') | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5CYL
 
 | |
4JCY
 
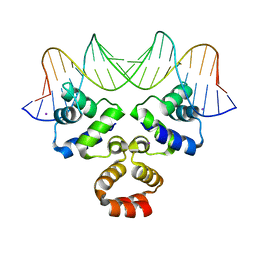 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OR operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*AP*AP*CP*TP*AP*AP*GP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*TP*AP*AP*GP*AP*TP*TP*TP*TP*CP*TP*TP*AP*GP*TP*TP*T)-3'), ... | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, Mcgeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3J9G
 
 | | Atomic model of the VipA/VipB, the type six secretion system contractile sheath of Vibrio cholerae from cryo-EM | | Descriptor: | VipA, VipB | | Authors: | Kudryashev, M, Wang, R.Y.-R, Brackmann, M, Scherer, S, Maier, T, Baker, D, DiMaio, F, Stahlberg, H, Egelman, E.H, Basler, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Type VI Secretion System Contractile Sheath.
Cell(Cambridge,Mass.), 160, 2015
|
|
