3A0B
 
 | | Crystal structure of Br-substituted Photosystem II complex | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Kawakami, K, Umena, Y, Kamiya, N, Shen, J.-R. | | Deposit date: | 2009-03-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1G5S
 
 | | CRYSTAL STRUCTURE OF HUMAN CYCLIN DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH THE INHIBITOR H717 | | Descriptor: | 2-[TRANS-(4-AMINOCYCLOHEXYL)AMINO]-6-(BENZYL-AMINO)-9-CYCLOPENTYLPURINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Dreyer, M.K, Borcherding, D.R, Dumont, J.A, Peet, N.P, Tsay, J.T, Wright, P.S, Bitonti, A.J, Shen, J, Kim, S.-H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of human cyclin-dependent kinase 2 in complex with the adenine-derived inhibitor H717.
J.Med.Chem., 44, 2001
|
|
1YQ4
 
 | | Avian respiratory complex ii with 3-nitropropionate and ubiquinone | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 3-NITROPROPANOIC ACID, Coenzyme Q10, ... | | Authors: | Huang, L, Sun, G, Cobessi, D, Wang, A, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
2H89
 
 | | Avian Respiratory Complex II with Malonate Bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
1Z4U
 
 | | hepatitis C virus NS5B RNA-dependent RNA polymerase complex with inhibitor PHA-00799585 | | Descriptor: | (2Z)-2-[(1-ADAMANTYLCARBONYL)AMINO]-3-[4-(2-BROMOPHENOXY)PHENYL]PROP-2-ENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pfefferkorn, J.A, Greene, M, Nugent, R, Gross, R.J, Mitchell, M.A, Finzel, B.C, Harris, M.S, Wells, P.A, Shelly, J.A, Anstadt, R. | | Deposit date: | 2005-03-16 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibitors of HCV NS5B polymerase. Part 2: Evaluation of the northern region of (2Z)-2-benzoylamino-3-(4-phenoxy-phenyl)-acrylic acid
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2H88
 
 | | Avian Mitochondrial Respiratory Complex II at 1.8 Angstrom Resolution | | Descriptor: | AZIDE ION, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
1YVF
 
 | | Hepatitis C virus NS5B RNA-dependent RNA polymerase complex with inhibitor PHA-00729145 | | Descriptor: | (2Z)-2-(BENZOYLAMINO)-3-[4-(2-BROMOPHENOXY)PHENYL]-2-PROPENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pfefferkorn, J.A, Greene, M.L, Nugent, R.A, Gross, R.J, Mitchell, M.A, Finzel, B.C, Harris, M.S, Wells, P.A, Shelly, J.A, Anstadt, R.A, Kilkuskie, R.E, Kopta, L.A, Schwende, F.J. | | Deposit date: | 2005-02-15 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitors of HCV NS5B polymerase. Part 1: Evaluation of the southern region of (2Z)-2-(benzoylamino)-3-(5-phenyl-2-furyl)acrylic acid.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2FBW
 
 | | Avian respiratory complex II with carboxin bound | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, AZIDE ION, ... | | Authors: | Huang, L.S, Sun, G, Cobessi, D, Wang, A.C, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-12-10 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme.
J.Biol.Chem., 281, 2006
|
|
3A0H
 
 | | Crystal structure of I-substituted Photosystem II complex | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Kawakami, K, Umena, Y, Kamiya, N, Shen, J.-R. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4AUA
 
 | | Liganded X-ray crystal structure of cyclin dependent kinase 6 (CDK6) | | Descriptor: | 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone, CYCLIN-DEPENDENT KINASE 6 | | Authors: | Cho, Y.S, Angove, H, Brain, C, Chen, C.H.T, Cheng, R, Chopra, R, Chung, K, Congreve, M, Dagostin, C, Davis, D, Feltell, R, Giraldes, J, Hiscock, S, Kim, S, Kovats, S, Lagu, B, Lewry, K, Loo, A, Lu, Y, Luzzio, M, Maniara, W, Mcmenamin, R, Mortenson, P, Benning, R, O'Reilly, M, Rees, D, Shen, J, Smith, T, Wang, Y, Williams, G, Woolford, A, Wrona, W, Xu, M, Yang, F, Howard, S. | | Deposit date: | 2012-05-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
3BM0
 
 | |
3CGY
 
 | | Crystal Structure of Salmonella Sensor Kinase PhoQ catalytic domain in complex with radicicol | | Descriptor: | RADICICOL, Virulence sensor histidine kinase phoQ | | Authors: | Guarnieri, M.T, Zhang, L, Shen, J, Zhao, R. | | Deposit date: | 2008-03-06 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Hsp90 inhibitor radicicol interacts with the ATP-binding pocket of bacterial sensor kinase PhoQ.
J.Mol.Biol., 379, 2008
|
|
3CGZ
 
 | |
3WU2
 
 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Umena, Y, Kawakami, K, Shen, J.R, Kamiya, N. | | Deposit date: | 2014-04-21 | | Release date: | 2014-09-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A
Nature, 473, 2011
|
|
3WG7
 
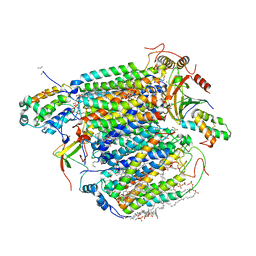 | | A 1.9 angstrom radiation damage free X-ray structure of large (420KDa) protein by femtosecond crystallography | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Hirata, K, Shinzawa-Itoh, K, Yano, N, Takemura, S, Kato, K, Hatanaka, M, Muramoto, K, Kawahara, T, Tsukihara, T, Yamashita, E, Tono, K, Ueno, G, Hikima, T, Murakami, H, Inubushi, Y, Yabashi, M, Ishikawa, T, Yamamoto, M, Ogura, T, Sugimoto, H, Shen, J.R, Yoshikawa, S, Ago, H. | | Deposit date: | 2013-07-29 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL.
Nat.Methods, 11, 2014
|
|
3J35
 
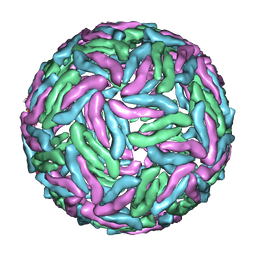 | | Cryo-EM reconstruction of Dengue virus at 37 C | | Descriptor: | envelope protein | | Authors: | Zhang, X.Z, Sheng, J, Plevka, P, Kuhn, R.J, Diamond, M.S, Rossmann, M.G. | | Deposit date: | 2013-02-24 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Dengue structure differs at the temperatures of its human and mosquito hosts.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1GML
 
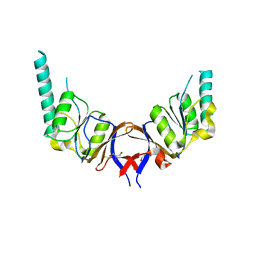 | | crystal structure of the mouse CCT gamma apical domain (triclinic) | | Descriptor: | GLYCEROL, T-COMPLEX PROTEIN 1 SUBUNIT GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-09-17 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
1GN1
 
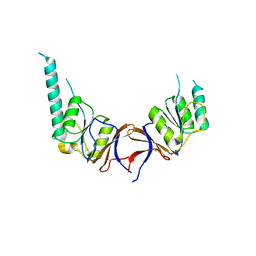 | | crystal structure of the mouse CCT gamma apical domain (monoclinic) | | Descriptor: | CALCIUM ION, CCT-GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-10-01 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
2A9H
 
 | | NMR structural studies of a potassium channel / charybdotoxin complex | | Descriptor: | Voltage-gated potassium channel, charybdotoxin | | Authors: | Yu, L, Sun, C, Song, D, Shen, J, Xu, N, Gunasekera, A, Hajduk, P.J, Olejniczak, E.T. | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-10 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies of a potassium channel-charybdotoxin complex.
Biochemistry, 44, 2005
|
|
1VCR
 
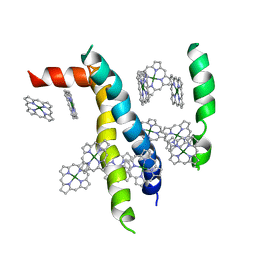 | | An icosahedral assembly of light-harvesting chlorophyll a/b protein complex from pea thylakoid membranes | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL B, Chlorophyll a-b binding protein AB80 | | Authors: | Hino, T, Kanamori, E, Shen, J.-R, Kouyama, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9.5 Å) | | Cite: | An icosahedral assembly of the light-harvesting chlorophyll a/b protein complex from pea chloroplast thylakoid membranes.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2GSW
 
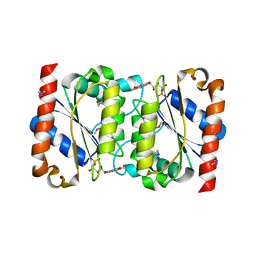 | | Crystal Structure of the Putative NADPH-dependent Azobenzene FMN-Reductase YhdA from Bacillus subtilis, Northeast Structural Genomics Target SR135 | | Descriptor: | FLAVIN MONONUCLEOTIDE, yhdA | | Authors: | Forouhar, F, Hussain, M, Jayaraman, S, Shen, J, Cooper, B, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal Structure of the Putative NADPH-dependent Azobenzene FMN-Reductase YhdA from Bacillus subtilis, Northeast Structural Genomics Target SR135
To be Published
|
|
1HZ2
 
 | | SOLUTION NMR STRUCTURE OF SELF-COMPLEMENTARY DUPLEX 5'-D(AGGCG*CCT)2 CONTAINING A TRIMETHYLENE CROSSLINK AT THE N2 POSITION OF G*. MODEL OF A MALONDIALDEHYDE CROSSLINK | | Descriptor: | DNA (5'-D(*AP*GP*GP*CP*GP*CP*CP*T)-3'), PROPANE | | Authors: | Dooley, P.A, Tsarouhtsis, D, Korbel, G.A, Nechev, L.V, Shearer, J, Zegar, I.S, Harris, C.M, Stone, M.P, Harris, T.M. | | Deposit date: | 2001-01-23 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of an oligodeoxynucleotide containing a trimethylene interstrand cross-link in a 5'-(CpG) motif: model of a malondialdehyde cross-link.
J.Am.Chem.Soc., 123, 2001
|
|
4DSI
 
 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA, Se-dGTP and Calcium | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA, ... | | Authors: | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | Deposit date: | 2012-02-19 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
4DWI
 
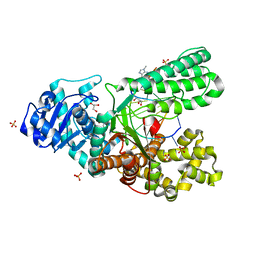 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with self complementary DNA, Se-dGTP and Calcium | | Descriptor: | 9-METHYLGUANINE, CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*AP*TP*CP*CP*A)-3'), ... | | Authors: | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | Deposit date: | 2012-02-24 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
4DSJ
 
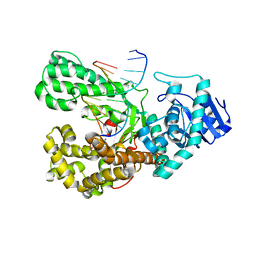 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA, dGTP and Calcium | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA, ... | | Authors: | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | Deposit date: | 2012-02-19 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
