5DWF
 
 | | Crystal structure of the complex of Peptidoglycan recognition protein, PGRP-S from camel with ethylene glycol at 1.83 A resolution | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Singh, P.K, Yadav, S.P, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-22 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the complex of Peptidoglycan recognition protein, PGRP-S from camel with ethylene glycol at 1.83 A resolution
To Be Published
|
|
5E0A
 
 | | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the complex of Camel Peptidoglycan Recognition Protein (CPGRP-S) and N-Acetylglucosamine at 2.6 A
To Be Published
|
|
9CYR
 
 | | Crystal structure of D245G mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYP
 
 | | Crystal structure of I19V mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYO
 
 | | Crystal structure of wild-type human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYQ
 
 | | Crystal structure of Q78R mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
3V14
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein complexed with Trehalose at 1.70 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein complexed with Trehalose at 1.70 A resolution
To be Published
|
|
3U6T
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein in complex with Kanamycin at 1.85 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, KANAMYCIN A, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-10-13 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein in complex with Kanamycin at 1.85 A
To be Published
|
|
3U8F
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein in complex with Mycolic acid at 1.8 A resolution | | Descriptor: | (2R,3R)-2-hexyl-3-hydroxytridecanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-10-17 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein in complex with Mycolic acid at 1.8 A resolution
To be Published
|
|
5ILW
 
 | | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with Uridine at 1.97 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
8I11
 
 | |
3TB2
 
 | | 1-Cys peroxidoxin from Plasmodium Yoelli | | Descriptor: | 1,2-ETHANEDIOL, 1-Cys peroxiredoxin, GLYCEROL, ... | | Authors: | Qiu, W, Artz, J.D, Vedadi, M, Sharma, S, Houston, S, Lew, J, Wasney, G, Amani, M, Xu, X, Bray, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Hui, R, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-08-04 | | Release date: | 2011-10-19 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
8IYN
 
 | |
8J68
 
 | |
5GH0
 
 | | Crystal structure of the complex of bovine lactoperoxidase with mercaptoimidazole at 2.3 A resolution | | Descriptor: | 1,3-dihydroimidazole-2-thione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Singh, A.K, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-06-17 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of anti-thyroid drugs: Binding studies and structure determination of the complex of lactoperoxidase with 2-mercaptoimidazole at 2.30 angstrom resolution
Proteins, 85, 2017
|
|
8IL9
 
 | |
5UNG
 
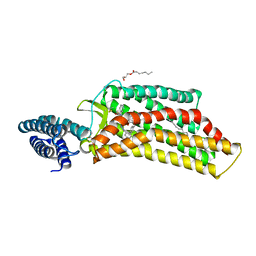 | | XFEL structure of human angiotensin II type 2 receptor (Orthorhombic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl] methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of Type-2 angiotensin II receptor and Soluble cytochrome b562, N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide, ... | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
5UNF
 
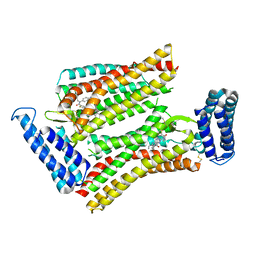 | | XFEL structure of human angiotensin II type 2 receptor (Monoclinic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]) | | Descriptor: | Chimera protein of Type-2 angiotensin II receptor and Soluble cytochrome b562, N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
5UNH
 
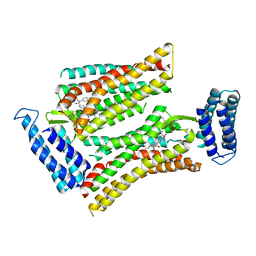 | | Synchrotron structure of human angiotensin II type 2 receptor in complex with compound 2 (N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl-3-{[2'-(2H-tetrazol-5-yl)[1,1'- biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide) | | Descriptor: | N-[(furan-2-yl)methyl]-N-(4-oxo-2-propyl-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)benzamide, Soluble cytochrome b562,Type-2 angiotensin II receptor | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
3TDF
 
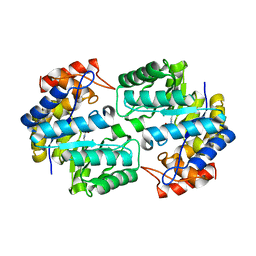 | | Crystal structure of the complex of Dihydrodipicolinate synthase from Acinetobacter baumannii with 2-Ketobutanoic acid at 1.99 A resolution | | Descriptor: | 2-KETOBUTYRIC ACID, Dihydrodipicolinate synthase | | Authors: | Kumar, M, Kaushik, S, Sinha, M, Kaur, P, Tewari, R, Sharma, S, Singh, T.P. | | Deposit date: | 2011-08-11 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the complex of Dihydrodipicolinate synthase from Acinetobacter baumannii with 2-Ketobutanoic acid at 1.99 A resolution
To be Published
|
|
8FU3
 
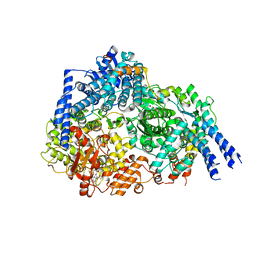 | | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | | Descriptor: | 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yu, X, Abeywickrema, P, Bonneux, B, Behera, I, Jacoby, E, Fung, A, Adhikary, S, Bhaumik, A, Carbajo, R.J, Bruyn, S.D, Miller, R, Patrick, A, Pham, Q, Piassek, M, Verheyen, N, Shareef, A, Sutto-Ortiz, P, Ysebaert, N, Vlijmen, H.V, Jonckers, T.H.M, Herschke, F, McLellan, J.S, Decroly, E, Fearns, R, Grosse, S, Roymans, D, Sharma, S, Rigaux, P, Jin, Z. | | Deposit date: | 2023-01-16 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor.
Commun Biol, 6, 2023
|
|
8FW2
 
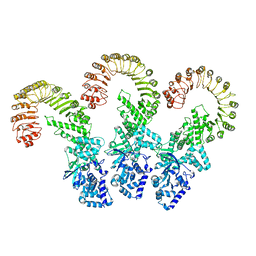 | | Cryo-EM structure of full-length human NLRC4 inflammasome with C11 symmetry | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Matico, R.E, Yu, X, Miller, R, Somani, S, Ricketts, M.D, Kumar, N, Steele, R.A, Medley, Q, Berger, S, Faustin, B, Sharma, S. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of the human NAIP/NLRC4 inflammasome assembly and pathogen sensing.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FVU
 
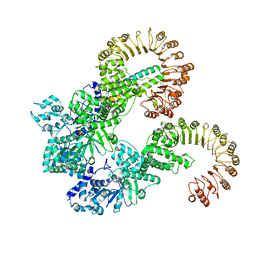 | | Cryo-EM structure of human Needle/NAIP/NLRC4 (R288A) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Baculoviral IAP repeat-containing protein 1, Lethal factor,Type III secretion system protein, ... | | Authors: | Matico, R.E, Yu, X, Miller, R, Somani, S, Ricketts, M.D, Kumar, N, Steele, R.A, Medley, Q, Berger, S, Faustin, B, Sharma, S. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of the human NAIP/NLRC4 inflammasome assembly and pathogen sensing.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FW9
 
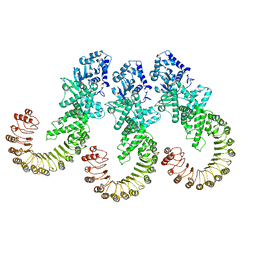 | | Cryo-EM structure of full-length human NLRC4 inflammasome with C12 symmetry | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Matico, R.E, Yu, X, Miller, R, Somani, S, Ricketts, M.D, Kumar, N, Steele, R.A, Medley, Q, Berger, S, Faustin, B, Sharma, S. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | Structural basis of the human NAIP/NLRC4 inflammasome assembly and pathogen sensing.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3S8H
 
 | | Structure of dihydrodipicolinate synthase complexed with 3-Hydroxypropanoic acid(HPA)at 2.70 A resolution | | Descriptor: | 3-HYDROXY-PROPANOIC ACID, Dihydrodipicolinate synthase | | Authors: | Kumar, M, Kaur, N, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-05-28 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of dihydrodipicolinate synthase complexed with 3-Hydroxypropanoic acid(HPA)at 2.70 A resolution
TO BE PUBLISHED
|
|
