4DN6
 
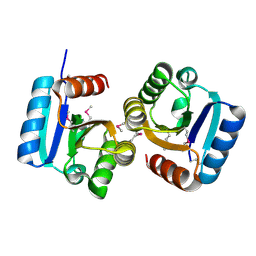 | |
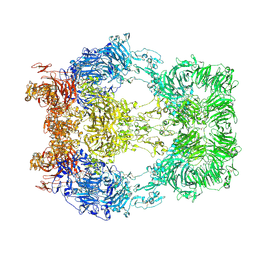
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
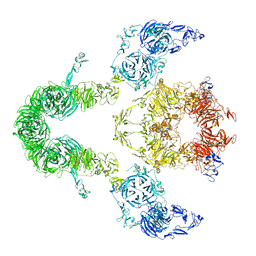
8EM7
 
 | | Cryo-EM structure of LRP2 at pH 5.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
8EM4
 
 | | Cryo-EM structure of LRP2 at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Brasch, J, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
2V0C
 
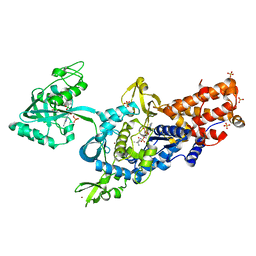 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A SULPHAMOYL ANALOGUE OF LEUCYL-ADENYLATE In the synthetic site and an adduct of AMP with 5-Fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole (AN2690) in the editing site | | Descriptor: | AMINOACYL-TRNA SYNTHETASE, LEUCINE, SULFATE ION, ... | | Authors: | Rock, F, Mao, W, Yaremchuk, A, Tukalo, M, Crepin, T, Zhou, H, Zhang, Y, Hernandez, V, Akama, T, Baker, S, Plattner, J, Shapiro, L, Martinis, S.A, Benkovic, S.J, Cusack, S, Alley, M.R.K. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Antifungal Agent Inhibits an Aminoacyl-tRNA Synthetase by Trapping tRNA in the Editing Site.
Science, 316, 2007
|
|
1I7E
 
 | | C-Terminal Domain Of Mouse Brain Tubby Protein bound to Phosphatidylinositol 4,5-bis-phosphate | | Descriptor: | L-ALPHA-GLYCEROPHOSPHO-D-MYO-INOSITOL-4,5-BIS-PHOSPHATE, TUBBY PROTEIN | | Authors: | Santagata, S, Boggon, T.J, Baird, C.L, Shan, W.S, Shapiro, L. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | G-protein signaling through tubby proteins.
Science, 292, 2001
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
2V0G
 
 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A tRNA(leu) transcript with 5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1- BENZOXABOROLE (AN2690) forming an adduct to the ribose of adenosine- 76 in the enzyme editing site. | | Descriptor: | AMINOACYL-TRNA SYNTHETASE, LEUCINE, MERCURY (II) ION, ... | | Authors: | Rock, F, Mao, W, Yaremchuk, A, Tukalo, M, Crepin, T, Zhou, H, Zhang, Y, Hernandez, V, Akama, T, Baker, S, Plattner, J, Shapiro, L, Martinis, S.A, Benkovic, S.J, Cusack, S, Alley, M.R.K. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An Antifungal Agent Inhibits an Aminoacyl-tRNA Synthetase by Trapping tRNA in the Editing Site.
Science, 316, 2007
|
|
5ZJT
 
 | | Structure of AbdB/Exd complex bound to a 'Black14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*AP*AP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*TP*TP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Zeiske, T, Baburajendran, N, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
5ZJS
 
 | | Structure of AbdB/Exd complex bound to a 'Blue14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*AP*TP*TP*AP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*TP*TP*AP*AP*TP*CP*AP*TP*GP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Baburajendran, N, Zeiske, T, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
7JGZ
 
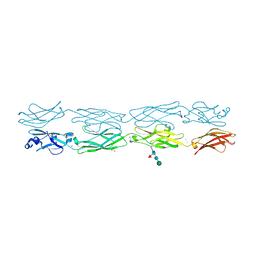 | | Protocadherin gammaC4 EC1-4 crystal structure | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition.
Elife, 11, 2022
|
|
5ZJR
 
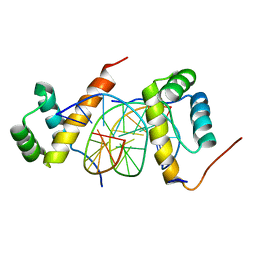 | | Structure of AbdB/Exd complex bound to a 'Magenta14' DNA sequence | | Descriptor: | DNA (5'-D(*GP*TP*CP*GP*TP*AP*AP*AP*TP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*AP*TP*TP*TP*AP*CP*GP*AP*C)-3'), Homeobox protein abdominal-B, ... | | Authors: | Zeiske, T, Baburajendran, N, Kaczynska, A, Mann, R, Honig, B, Shapiro, L, Palmer, A.G. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites.
Cell Rep, 24, 2018
|
|
2QRC
 
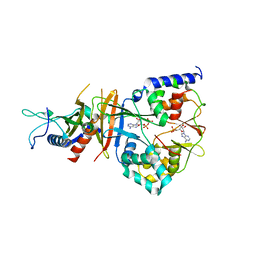 | | Crystal structure of the adenylate sensor from AMP-activated protein kinase in complex with ADP and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Protein C1556.08c, ... | | Authors: | Jin, X, Townley, R, Shapiro, L. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into AMPK Regulation: ADP Comes into Play.
Structure, 15, 2007
|
|
5K6X
 
 | | Sidekick-2 immunoglobulin domains 1-4, crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
5K6Y
 
 | | Sidekick-2 immunoglobulin domains 1-4, crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Protein sidekick-2 | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
6EG0
 
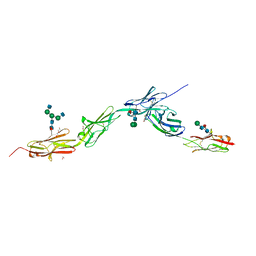 | | Crystal structure of Dpr4 Ig1-Ig2 in complex with DIP-Eta Ig1-Ig3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cosmanescu, F, Shapiro, L. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
6EFY
 
 | | Crystal Structure of DIP-Alpha Ig1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Dpr-interacting protein alpha, isoform A, ... | | Authors: | Cosmanescu, F, Shapiro, L. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
6EFZ
 
 | | Crystal Structure of DIP-Theta Ig1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cosmanescu, F, Shapiro, L. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
6EG1
 
 | | Crystal structure of Dpr2 Ig1-Ig2 in complex with DIP-Theta Ig1-Ig3 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cosmanescu, F, Patel, S, Shapiro, L. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins.
Neuron, 100, 2018
|
|
8DXS
 
 | |
8DW9
 
 | |
8DWA
 
 | |
7RGF
 
 | | Protocadherin gammaC4 EC1-4 crystal structure disrupted trans interface | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition.
Elife, 11, 2022
|
|
2A62
 
 | | Crystal structure of mouse cadherin-8 EC1-3 | | Descriptor: | CALCIUM ION, Cadherin-8 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Schieren, I, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-07-01 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
2AKO
 
 | |
