6FMY
 
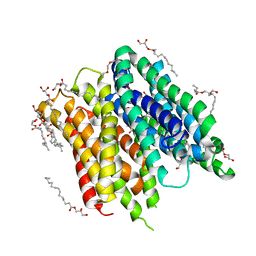 | | IMISX-EP of S-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Di-or tripeptide:H+ symporter, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
3MGV
 
 | | Cre recombinase-DNA transition state | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*G)-3'), Recombinase cre, ... | | Authors: | Gibb, B.P, Gupta, K, Ghosh, K, Sharp, R, Chen, J, Van Duyne, G.D. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Requirements for catalysis in the Cre recombinase active site.
Nucleic Acids Res., 38, 2010
|
|
2L0B
 
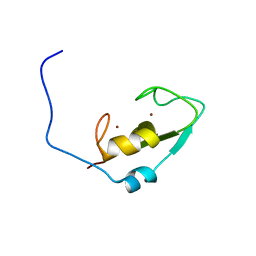 | | Solution NMR structure of zinc finger domain of E3 ubiquitin-protein ligase praja-1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) target HR4710B | | Descriptor: | E3 ubiquitin-protein ligase Praja-1, ZINC ION | | Authors: | Liu, G, Tong, S, Hamilton, K, Ciccosanti, C, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of zinc finger domain of E3 ubiquitin-protein ligase protein praja-1 from Homo sapiens, northeast structural genomics consortium (NESG) target HR4710B
To be Published
|
|
2L6U
 
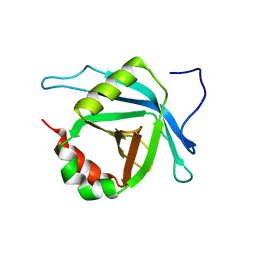 | | Solution NMR Structure of Med25(391-543) Comprising the Activator-Interacting Domain (ACID) of Human Mediator Subuniti 25. Northeast Structural Genomics Consortium Target HR6188A | | Descriptor: | Mediator complex subunit MED25 | | Authors: | Eletsky, A, Ryuechan, W.T, Sukumaran, D.K, Shastry, R, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of MED25(391-543) comprising the activator-interacting domain (ACID) of human mediator subunit 25.
J.Struct.Funct.Genom., 12, 2011
|
|
2LNA
 
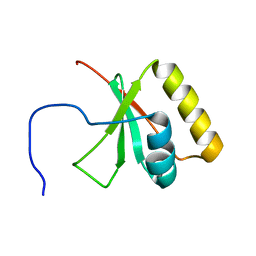 | | Solution NMR Structure of the mitochondrial inner membrane domain (residues 164-251), FtsH_ext, from the paraplegin-like protein AFG3L2 from Homo sapiens, Northeast Structural Genomics Consortium Target HR6741A | | Descriptor: | AFG3-like protein 2 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Janua, H, Kohan, E, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and MD simulations of the AAA protease intermembrane space domain indicates peripheral membrane localization within the hexaoligomer.
Febs Lett., 587, 2013
|
|
2KKQ
 
 | | Solution NMR Structure of the Ig-like C2-type 2 Domain of Human Myotilin. Northeast Structural Genomics Target HR3158. | | Descriptor: | Myotilin | | Authors: | Rossi, P, Shastry, R, Ciccosanti, C, Hamilton, K, Xiao, R, Acton, T.B, Swapna, G.V.T, Nair, R, Everett, J.K, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Ig-like C2-type 2 Domain of Human Myotilin. Northeast Structural Genomics Target HR3158.
To be Published
|
|
2JWZ
 
 | | Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins | | Descriptor: | Ubiquitin | | Authors: | Haririnia, A, Verma, R, Purohit, N, Twarog, M, Deshaies, R, Bolon, D, Fushman, D. | | Deposit date: | 2007-10-31 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins.
J.Mol.Biol., 375, 2008
|
|
1YVW
 
 | | Crystal structure of Phosphoribosyl-ATP pyrophosphohydrolase from Bacillus cereus. NESGC target BcR13. | | Descriptor: | Phosphoribosyl-ATP pyrophosphatase | | Authors: | Benach, J, Kuzin, A.P, Forouhar, F, Abashidze, M, Vorobiev, S.M, Shastry, R, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Phosphoribosyl-ATP pyrophosphohydrolase from Bacillus cereus at 2.6 A resolution.
To be Published
|
|
4E68
 
 | | Unphosphorylated STAT3B core protein binding to dsDNA | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), Signal transducer and activator of transcription 3 | | Authors: | Collie, G.W, Parkinson, G.N, Shah, R. | | Deposit date: | 2012-03-15 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Observation of unphosphorylated STAT3 core protein binding to target dsDNA by PEMSA and X-ray crystallography.
Febs Lett., 587, 2013
|
|
1XFS
 
 | | X-Ray Crystal Structure of Protein NE0264 from Nitrosomonas europaea. Northeast Structural Genomics Consortium Target NeR5. | | Descriptor: | conserved hypothetical protein | | Authors: | Forouhar, F, Abashidze, M, Vorobiev, S.M, Ciano, M, Ma, L.-C, Shastry, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-15 | | Release date: | 2004-09-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the hypothetical Protein from Nitrosomonas europaea, NESG Target NeR5
To be Published
|
|
2AEG
 
 | | X-Ray Crystal Structure of Protein Atu5096 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR63. | | Descriptor: | hypothetical protein AGR_pAT_140 | | Authors: | Forouhar, F, Abashidze, M, Kuzin, A.P, Vorobiev, S.M, Shastry, R, Cooper, B, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-07-22 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Hypothetical Protein Atu5096 from Agrobacterium tumefaciens.
To be Published
|
|
1NY8
 
 | | Solution structure of Protein yrbA from Escherichia Coli: Northeast Structural Genomics Consortium target ER115 | | Descriptor: | Protein yrbA | | Authors: | Swapna, G.V.T, Huang, J.Y, Acton, T.B, Shastry, R, Chiang, Y.-W, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2004-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Protein yrbA from Escherichia Coli: Northeast Structural Genomics Consortium target ER115
To be Published
|
|
2K1H
 
 | | Solution NMR structure of SeR13 from Staphylococcus epidermidis. Northeast Structural Genomics Consortium target SeR13 | | Descriptor: | Uncharacterized protein SeR13 | | Authors: | Lee, H, Wylie, G, Bansal, S, Wang, X, Shastry, R, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of SeR13.
To be Published
|
|
2JWH
 
 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
2LCE
 
 | | Chemical shift assignment of Hr4436B from Homo Sapiens, Northeast Structural Genomics Consortium | | Descriptor: | B-cell lymphoma 6 protein, ZINC ION | | Authors: | Lee, H, Shastry, R, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Hr4436B.
To be Published
|
|
2JWG
 
 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
2M9A
 
 | | Solution NMR Structure of E3 ubiquitin-protein ligase ZFP91 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7784A | | Descriptor: | E3 ubiquitin-protein ligase ZFP91, ZINC ION | | Authors: | Pederson, K, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Hr7784A
To be Published
|
|
2M1L
 
 | | Solution NMR Structure of Cyclin-dependent kinase 2-associated protein 2 (CDK2AP2, DOC-1R) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8910C | | Descriptor: | Cyclin-dependent kinase 2-associated protein 2 | | Authors: | Ertekin, A, Janjua, H, Kohan, E, Shastry, R, Pederson, K, Prestegard, J.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of CDK2-associated protein 2 (CDK2AP2, Deleted in Oral Cancer 1 Related protein, DOC-1R)
To be Published
|
|
8EDO
 
 | |
8EDN
 
 | |
8EDL
 
 | |
2MYI
 
 | | Solution Structure of Crc from P. syringae Lz4W | | Descriptor: | Exodeoxyribonuclease III | | Authors: | Deshmukh, M.V, Sharma, R. | | Deposit date: | 2015-01-25 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crc of Pseudomonas syringae Lz4W utilizes the dominant RNA binding site for mutually exclusive interactions with Hfq:mRNA and CrcY/Z RNA
J.Magn.Reson., 10-11, 2022
|
|
6AVK
 
 | | Streptavidin bound to peptide-like compound KPM-6 | | Descriptor: | 1-hydroxydodecan-4-one, N-[(2H-1,3-benzodioxol-5-yl)methyl]-2-({[(2H-1,3-benzodioxol-5-yl)methyl][2-(chloromethyl)-1,3-oxazole-4-carbonyl]amino}methyl)-N-[(4-carbamoyl-1,3-oxazol-2-yl)methyl]-1,3-oxazole-4-carboxamide, Streptavidin | | Authors: | Park, H, Shamim, R, McEnaney, P, Kodadek, T. | | Deposit date: | 2017-09-02 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Efficient Workflow for Screening DNA-encoded One Bead One Compound Libraries Using a Flow Cytometer
To be Published
|
|
4GRV
 
 | | The crystal structure of the neurotensin receptor NTS1 in complex with neurotensin (8-13) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Neurotensin 8-13, Neurotensin receptor type 1, ... | | Authors: | Noinaj, N, White, J.F, Shibata, Y, Love, J, Kloss, B, Xu, F, Gvozdenovic-Jeremic, J, Shah, P, Shiloach, J, Tate, C.G, Grisshammer, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the agonist-bound neurotensin receptor.
Nature, 490, 2012
|
|
3CER
 
 | | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13 | | Descriptor: | Possible exopolyphosphatase-like protein, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Chen, Y, Neely, H, Seetharaman, J, Shastry, R, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-04-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13.
To be Published
|
|
