2LNA
 
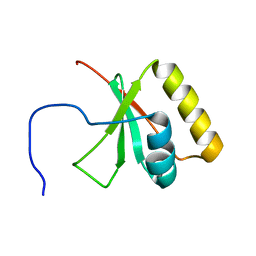 | | Solution NMR Structure of the mitochondrial inner membrane domain (residues 164-251), FtsH_ext, from the paraplegin-like protein AFG3L2 from Homo sapiens, Northeast Structural Genomics Consortium Target HR6741A | | Descriptor: | AFG3-like protein 2 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Janua, H, Kohan, E, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and MD simulations of the AAA protease intermembrane space domain indicates peripheral membrane localization within the hexaoligomer.
Febs Lett., 587, 2013
|
|
3FB2
 
 | | Crystal structure of the human brain alpha spectrin repeats 15 and 16. Northeast Structural Genomics Consortium target HR5563a. | | Descriptor: | Spectrin alpha chain, brain spectrin | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Shastry, R, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the human brain alpha spectrin repeats 15 and 16.
To be Published
|
|
3EXA
 
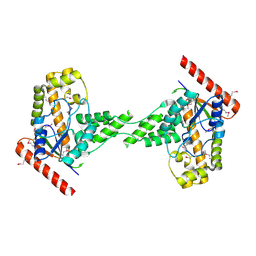 | | Crystal structure of the full-length tRNA isopentenylpyrophosphate transferase (BH2366) from Bacillus halodurans, Northeast Structural Genomics Consortium target BhR41. | | Descriptor: | tRNA delta(2)-isopentenylpyrophosphate transferase | | Authors: | Forouhar, F, Abashidze, M, Neely, H, Seetharaman, J, Shastry, R, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the full-length tRNA isopentenylpyrophosphate transferase (BH2366) from Bacillus halodurans, Northeast Structural Genomics Consortium target BhR41.
To be Published
|
|
2L33
 
 | | Solution NMR Structure of DRBM 2 domain of Interleukin enhancer-binding factor 3 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4527E | | Descriptor: | Interleukin enhancer-binding factor 3 | | Authors: | Liu, G, Janjua, H, Xiao, R, Acton, T.B, Ciccosanti, A, Shastry, R.B, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-03 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR4527E
To be Published
|
|
2L6U
 
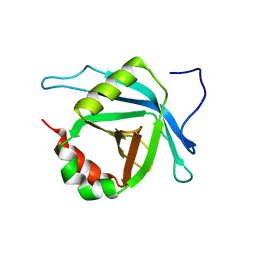 | | Solution NMR Structure of Med25(391-543) Comprising the Activator-Interacting Domain (ACID) of Human Mediator Subuniti 25. Northeast Structural Genomics Consortium Target HR6188A | | Descriptor: | Mediator complex subunit MED25 | | Authors: | Eletsky, A, Ryuechan, W.T, Sukumaran, D.K, Shastry, R, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of MED25(391-543) comprising the activator-interacting domain (ACID) of human mediator subunit 25.
J.Struct.Funct.Genom., 12, 2011
|
|
1ERD
 
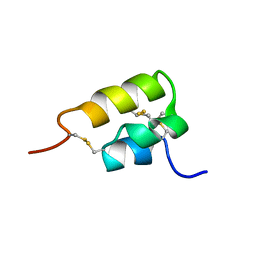 | | THE NMR SOLUTION STRUCTURE OF THE PHEROMONE ER-2 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI | | Descriptor: | PHEROMONE ER-2 | | Authors: | Ottiger, M, Szyperski, T, Luginbuhl, P, Ortenzi, C, Luporini, P, Bradshaw, R.A, Wuthrich, K. | | Deposit date: | 1994-02-14 | | Release date: | 1994-10-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-2 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 3, 1994
|
|
7LK3
 
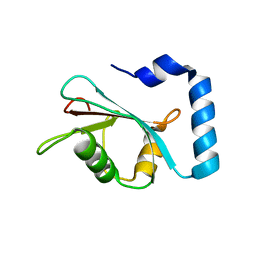 | | Crystal structure of untwinned human GABARAPL2 | | Descriptor: | 1,2-ETHANEDIOL, Gamma-aminobutyric acid receptor-associated protein-like 2 | | Authors: | Scicluna, K, Dewson, G, Czabotar, P.E, Birkinshaw, R.W. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new crystal form of GABARAPL2.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
5E0O
 
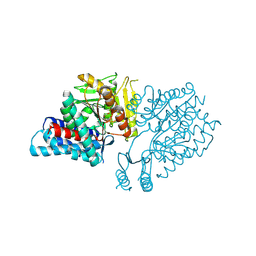 | |
6DNW
 
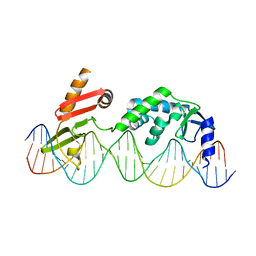 | | Sequence Requirements of the Listeria innocua prophage attP site | | Descriptor: | DNA (26-MER), Putative integrase [Bacteriophage A118], ZINC ION | | Authors: | Li, H, Sharp, R, Rutherford, K, Gupta, K, Van Duyne, G.D. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Serine Integrase attP Binding and Specificity.
J. Mol. Biol., 430, 2018
|
|
1NXU
 
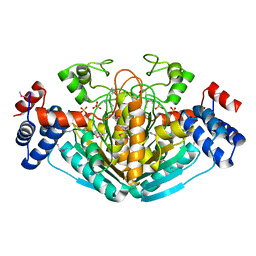 | | CRYSTAL STRUCTURE OF E. COLI HYPOTHETICAL OXIDOREDUCTASE YIAK NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82. | | Descriptor: | Hypothetical oxidoreductase yiaK, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Shastry, R, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
2L0B
 
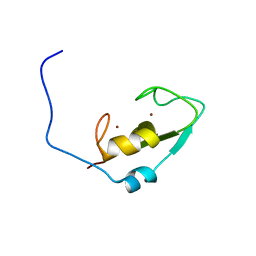 | | Solution NMR structure of zinc finger domain of E3 ubiquitin-protein ligase praja-1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) target HR4710B | | Descriptor: | E3 ubiquitin-protein ligase Praja-1, ZINC ION | | Authors: | Liu, G, Tong, S, Hamilton, K, Ciccosanti, C, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of zinc finger domain of E3 ubiquitin-protein ligase protein praja-1 from Homo sapiens, northeast structural genomics consortium (NESG) target HR4710B
To be Published
|
|
1NY7
 
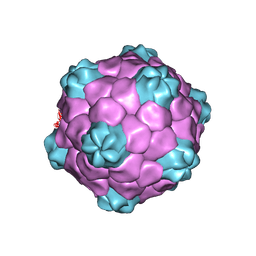 | | COWPEA MOSAIC VIRUS (CPMV) | | Descriptor: | COWPEA MOSAIC VIRUS, LARGE (L) SUBUNIT, SMALL (S) SUBUNIT | | Authors: | Lin, T, Chen, Z, Usha, R, Stauffacher, C.V, Dai, J.-B, Schmidt, T, Johnson, J.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Refined Crystal Structure of Cowpea Mosaic Virus at 2.8A Resolution
Virology, 265, 1999
|
|
2M4Q
 
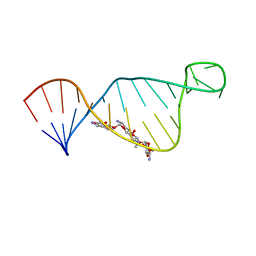 | | NMR structure of E. coli ribosomela decoding site with apramycin | | Descriptor: | APRAMYCIN, RNA (27-MER) | | Authors: | Puglisi, J.D, Tsai, A, Marshall, R, Viani, E. | | Deposit date: | 2013-02-10 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The impact of aminoglycosides on the dynamics of translation elongation.
Cell Rep, 3, 2013
|
|
1LQG
 
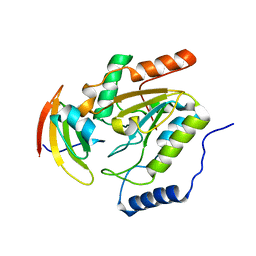 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1YF8
 
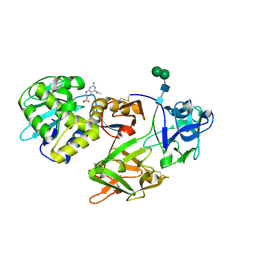 | | Crystal structure of Himalayan mistletoe RIP reveals the presence of a natural inhibitor and a new functionally active sugar-binding site | | Descriptor: | 2-AMINO-4-ISOPROPYL-PTERIDINE-6-CARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mishra, V, Bilgrami, S, Sharma, R.S, Kaur, P, Yadav, S, Betzel, C, Babu, C.R, Singh, T.P. | | Deposit date: | 2004-12-31 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of himalayan mistletoe ribosome-inactivating protein reveals the presence of a natural inhibitor and a new functionally active sugar-binding site.
J.Biol.Chem., 280, 2005
|
|
2AEG
 
 | | X-Ray Crystal Structure of Protein Atu5096 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR63. | | Descriptor: | hypothetical protein AGR_pAT_140 | | Authors: | Forouhar, F, Abashidze, M, Kuzin, A.P, Vorobiev, S.M, Shastry, R, Cooper, B, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-07-22 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Hypothetical Protein Atu5096 from Agrobacterium tumefaciens.
To be Published
|
|
1LQJ
 
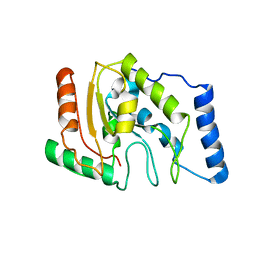 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6BNH
 
 | | Solution NMR structures of BRD4 ET domain with JMJD6 peptide | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, Bromodomain-containing protein 4 | | Authors: | Konuma, T, Yu, D, Zhao, C, Ju, Y, Sharma, R, Ren, C, Zhang, Q, Zhou, M.-M, Zeng, L. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Sci Rep, 7, 2017
|
|
5D52
 
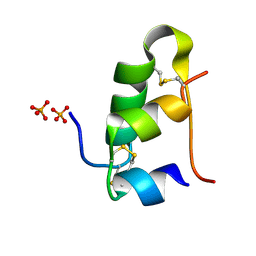 | | In meso in situ serial X-ray crystallography structure of insulin at room temperature | | Descriptor: | Insulin A chain, Insulin B chain, PHOSPHATE ION | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5E
 
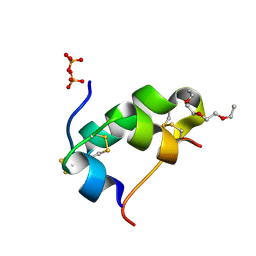 | | In meso in situ serial X-ray crystallography structure of insulin by sulfur-SAD at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Insulin A chain, Insulin B chain, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2LCE
 
 | | Chemical shift assignment of Hr4436B from Homo Sapiens, Northeast Structural Genomics Consortium | | Descriptor: | B-cell lymphoma 6 protein, ZINC ION | | Authors: | Lee, H, Shastry, R, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Hr4436B.
To be Published
|
|
1TIY
 
 | | X-RAY STRUCTURE OF GUANINE DEAMINASE FROM BACILLUS SUBTILIS NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR160 | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Kuzin, A.P, Vorobiev, S, Edstrom, W, Forouhar, F, Acton, T, Shastry, R, Ma, L.-C, Chiang, Y.-W, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-RAY STRUCTURE OF GUANINE DEAMINASE FROM BACILLUS SUBTILIS
NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR160
To be published
|
|
2M1L
 
 | | Solution NMR Structure of Cyclin-dependent kinase 2-associated protein 2 (CDK2AP2, DOC-1R) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8910C | | Descriptor: | Cyclin-dependent kinase 2-associated protein 2 | | Authors: | Ertekin, A, Janjua, H, Kohan, E, Shastry, R, Pederson, K, Prestegard, J.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of CDK2-associated protein 2 (CDK2AP2, Deleted in Oral Cancer 1 Related protein, DOC-1R)
To be Published
|
|
4KSR
 
 | | Crystal Structure of the Vibrio cholerae ATPase GspE Hexamer | | Descriptor: | Type II secretion system protein E, Hemolysin-coregulated protein | | Authors: | Hol, W.G, Turley, S, Lu, C.Y, Park, Y.J, Marionni, S.T, Lee, K, Patrick, M, Sandkvist, M, Bush, M, Shah, R. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Hexamers of the Type II Secretion ATPase GspE from Vibrio cholerae with Increased ATPase Activity.
Structure, 21, 2013
|
|
1R5X
 
 | |
