4ZR1
 
 | | Hydroxylase domain of scs7p | | 分子名称: | Ceramide very long chain fatty acid hydroxylase SCS7, TRIDECANE, ZINC ION, ... | | 著者 | Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G, Membrane Protein Structural Biology Consortium (MPSBC) | | 登録日 | 2015-05-11 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.6008 Å) | | 主引用文献 | The Crystal Structure of an Integral Membrane Fatty Acid alpha-Hydroxylase.
J.Biol.Chem., 290, 2015
|
|
4ZR0
 
 | |
5XNY
 
 | | Crystal structure of CreD | | 分子名称: | CreD | | 著者 | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | 登録日 | 2017-05-25 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
6F6Y
 
 | |
7E4D
 
 | | Crystal structure of PlDBR | | 分子名称: | Double Bond Reductase | | 著者 | Sugimoto, K, Senda, M, Senda, T. | | 登録日 | 2021-02-11 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Exploration and structure-based engineering of alkenal double bond reductases catalyzing the C alpha C beta double bond reduction of coniferaldehyde.
N Biotechnol, 68, 2022
|
|
3N23
 
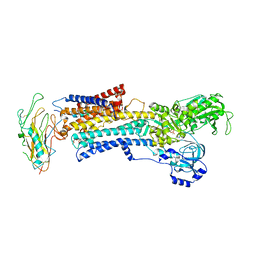 | | Crystal structure of the high affinity complex between ouabain and the E2P form of the sodium-potassium pump | | 分子名称: | MAGNESIUM ION, Na+/K+ ATPase gamma subunit transcript variant a, OUABAIN, ... | | 著者 | Yatime, L, Laursen, M, Morth, J.P, Esmann, M, Nissen, P, Fedosova, N.U. | | 登録日 | 2010-05-17 | | 公開日 | 2011-01-19 | | 最終更新日 | 2014-09-17 | | 実験手法 | X-RAY DIFFRACTION (4.6 Å) | | 主引用文献 | Structural insights into the high affinity binding of cardiotonic steroids to the Na+,K+-ATPase.
J.Struct.Biol., 174, 2011
|
|
4H9G
 
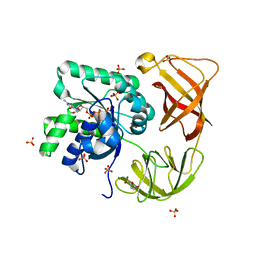 | | Probing EF-Tu with a very small brominated fragment library identifies the CCA pocket | | 分子名称: | 5-bromofuran-2-carboxylic acid, AMMONIUM ION, Elongation factor Tu-A, ... | | 著者 | Groftehauge, M.K, Therkelsen, M, Taaning, R.H, Skrydstrup, T, Morth, J.P, Nissen, P. | | 登録日 | 2012-09-24 | | 公開日 | 2013-09-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4HHR
 
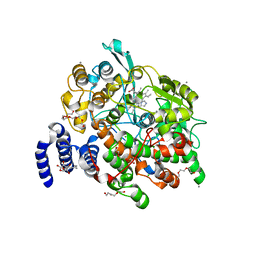 | | Crystal Structure of fatty acid alpha-dioxygenase (Arabidopsis thaliana) | | 分子名称: | Alpha-dioxygenase, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Goulah, C.C, Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | 登録日 | 2012-10-10 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | The crystal structure of alpha-Dioxygenase provides insight into diversity in the cyclooxygenase-peroxidase superfamily.
Biochemistry, 52, 2013
|
|
3L9J
 
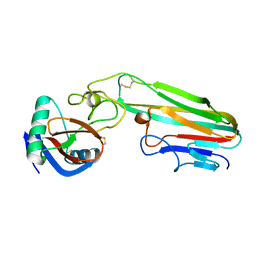 | | Selection of a novel highly specific TNFalpha antagonist: Insight from the crystal structure of the antagonist-TNFalpha complex | | 分子名称: | MAGNESIUM ION, TNFalpha, Tumor necrosis factor, ... | | 著者 | Byla, P, Andersen, M.H, Thogersen, H.C, Gad, H.H, Hartmann, R. | | 登録日 | 2010-01-05 | | 公開日 | 2010-02-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Selection of a novel and highly specific TNF{alpha} antagonist: insight from the crystal structure of the antagonist-TNF{alpha} complex
J.Biol.Chem., 285, 2010
|
|
6ISV
 
 | | Structure of acetophenone reductase from Geotrichum candidum NBRC 4597 in complex with NAD | | 分子名称: | Acetophenone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | 著者 | Koesoema, A.A, Sugiyama, Y, Senda, M, Senda, T, Matsuda, T. | | 登録日 | 2018-11-19 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for a highly (S)-enantioselective reductase towards aliphatic ketones with only one carbon difference between side chain.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
6K4H
 
 | |
6TRI
 
 | | CI-MOR repressor-antirepressor complex of the temperate bacteriophage TP901-1 from Lactococcus lactis | | 分子名称: | CI, MOR, SULFATE ION | | 著者 | Rasmussen, K.K, Blackledge, M, Herrmann, T, Jensen, M.R, Lo Leggio, L. | | 登録日 | 2019-12-18 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.277 Å) | | 主引用文献 | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4IL3
 
 | | Crystal Structure of S. mikatae Ste24p | | 分子名称: | Ste24p, ZINC ION | | 著者 | Pryor Jr, E.E, Horanyi, P.S, Clark, K, Fedoriw, N, Connelly, S.M, Koszelak-Rosenblum, M, Zhu, G, Malkowski, M.G, Dumont, M.E, Wiener, M.C, Membrane Protein Structural Biology Consortium (MPSBC) | | 登録日 | 2012-12-28 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Structure of the integral membrane protein CAAX protease Ste24p.
Science, 339, 2013
|
|
6GJE
 
 | | Structure of the Amnionless(20-357)-Cubilin(36-135) complex | | 分子名称: | Cubilin, Protein amnionless | | 著者 | Larsen, C, Etzerodt, A, Madsen, M, Skjoedt, K, Moestrup, S.K, Andersen, C.B.F. | | 登録日 | 2018-05-16 | | 公開日 | 2018-12-19 | | 最終更新日 | 2019-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural assembly of the megadalton-sized receptor for intestinal vitamin B12uptake and kidney protein reabsorption.
Nat Commun, 9, 2018
|
|
6K4G
 
 | |
6ZMV
 
 | | Structure of muramidase from Trichobolus zukalii | | 分子名称: | GLYCEROL, SULFATE ION, muramidase | | 著者 | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | 登録日 | 2020-07-04 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
5MM9
 
 | | VIM-2_2b. Metallo-beta-Lactamase Inhibitors by Bioisosteric Replacement: Preparation, Activity and Binding | | 分子名称: | (2~{R})-2-diethoxyphosphoryl-5-phenyl-pentane-1-thiol, MAGNESIUM ION, Metallo-beta-lactamase VIM-17, ... | | 著者 | Skagseth, S, Akhter, S, Paulsen, M.H, Samuelsen, O, Muhammad, Z, Leiros, H.-K.S, Bayer, A. | | 登録日 | 2016-12-08 | | 公開日 | 2017-03-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Metallo-beta-lactamase inhibitors by bioisosteric replacement: Preparation, activity and binding.
Eur J Med Chem, 135, 2017
|
|
6VJT
 
 | |
7AJ0
 
 | | Crystal structure of PsFucS1 sulfatase from Pseudoalteromonas sp. | | 分子名称: | Arylsulfatase, CALCIUM ION, CHLORIDE ION | | 著者 | Roret, T, Mikkelsen, M.D, Czjzek, M, Meyer, A.S. | | 登録日 | 2020-09-28 | | 公開日 | 2021-09-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A novel thermostable prokaryotic fucoidan active sulfatase PsFucS1 with an unusual quaternary hexameric structure.
Sci Rep, 11, 2021
|
|
6VR0
 
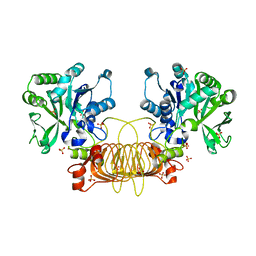 | | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A | | 分子名称: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | 著者 | Mascarenhas, R.N, Liu, D, Ballicora, M, Iglesias, A, Asencion, M, Figueroa, C. | | 登録日 | 2020-02-06 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A
To Be Published
|
|
6WO0
 
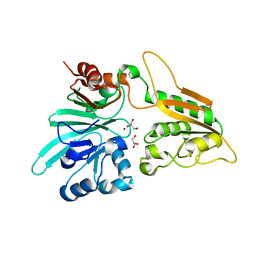 | | human Artemis/SNM1C catalytic domain, crystal form 1 | | 分子名称: | GLYCEROL, Protein artemis, ZINC ION | | 著者 | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | 登録日 | 2020-04-23 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
8PXL
 
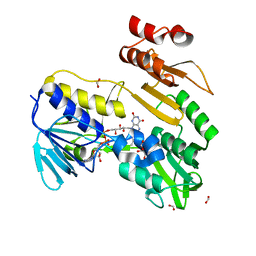 | | Structure of NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4, solved at wavelength 1.37 A | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, Ferredoxin reductase, ... | | 著者 | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Senda, M, Matsugaki, N, Kawano, Y, Wagner, A. | | 登録日 | 2023-07-23 | | 公開日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXK
 
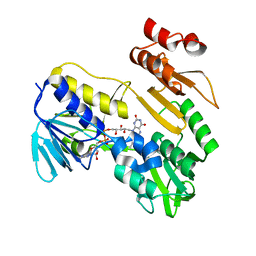 | | Structure of NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4, solved at wavelength 5.76 A | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin reductase | | 著者 | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Senda, M, Matsugaki, N, Kawano, Y, Wagner, A. | | 登録日 | 2023-07-23 | | 公開日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.77 Å) | | 主引用文献 | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
6WNL
 
 | | human Artemis/SNM1C catalytic domain, crystal form 2 | | 分子名称: | Protein artemis, ZINC ION | | 著者 | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | 登録日 | 2020-04-22 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
3K8G
 
 | | Structure of crystal form I of TP0453 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 30kLP | | 著者 | Zhu, G, Luthra, A, Desrosiers, D, Koszelak-Rosenblum, M, Mulay, V, Radolf, J.D, Malkowski, M.G. | | 登録日 | 2009-10-14 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Transition from Closed to Open Conformation of Treponema pallidum Outer Membrane-associated Lipoprotein TP0453 Involves Membrane Sensing and Integration by Two Amphipathic Helices.
J.Biol.Chem., 286, 2011
|
|
