5EPE
 
 | | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase, SODIUM ION | | Authors: | LaRowe, C, Shabalin, I.G, Kutner, J, Handing, K.B, Stead, M, Hillerich, B.S, Ahmed, M, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-11-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine
to be published
|
|
4WBT
 
 | | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Shabalin, I.G, Bacal, P, Kowalska, A.K, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate
to be published
|
|
4WJI
 
 | | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP and tyrosine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shabalin, I.G, Cooper, D.R, Hou, J, Zimmerman, M.D, Stead, M, Hillerich, B.S, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-30 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP
to be published
|
|
4XK2
 
 | | Crystal structure of aldo-keto reductase from Polaromonas sp. JS666 | | Descriptor: | Aldo/keto reductase, CHLORIDE ION, SODIUM ION | | Authors: | Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Sroka, P, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-01-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aldo-keto reductase from Polaromonas sp. JS666
to be published
|
|
4XCV
 
 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, NADP-dependent 2-hydroxyacid dehydrogenase, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obadi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-18 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH
to be published
|
|
4XFR
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Bordetella bronchiseptica (BB3215), Target EFI-511620, with bound citrate, domain swapped dimer, space group P6522 | | Descriptor: | CITRIC ACID, Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-28 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XGJ
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Pectobacterium atrosepticum (ECA3761), Target EFI-511609, APO structure, domain swapped dimer | | Descriptor: | Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-30 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XFM
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Pectobacterium atrosepticum (ECA3761), Target EFI-511609, with bound D-threonate, domain swapped dimer | | Descriptor: | THREONATE ION, Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-27 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XG0
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Bordetella bronchiseptica (BB3215), Target EFI-511620, with bound citrate, domain swapped dimer, space group C2221 | | Descriptor: | CHLORIDE ION, CITRIC ACID, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-30 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4DUP
 
 | | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42 | | Descriptor: | quinone oxidoreductase | | Authors: | Kumaran, D, Rice, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
4DLL
 
 | | Crystal structure of a 2-hydroxy-3-oxopropionate reductase from Polaromonas sp. JS666 | | Descriptor: | 2-hydroxy-3-oxopropionate reductase, SULFATE ION | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of a 2-hydroxy-3-oxopropionate reductase from Polaromonas sp. JS666
To be Published
|
|
4QB5
 
 | | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118 | | Descriptor: | 1,2-ETHANEDIOL, Glyoxalase/bleomycin resistance protein/dioxygenase, SULFATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-06 | | Release date: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a glyoxalase/bleomycin resistance protein from Albidiferax ferrireducens T118
To be Published
|
|
4YYC
 
 | | Crystal structure of trimethylamine methyltransferase from Sinorhizobium meliloti in complex with unknown ligand | | Descriptor: | CHLORIDE ION, Putative trimethylamine methyltransferase, UNKNOWN LIGAND | | Authors: | Shabalin, I.G, Porebski, P.J, Gasiorowska, O.A, Handing, K.B, Niedzialkowska, E, Cymborowski, M.T, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
4MYM
 
 | | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, AL Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-27 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a glyoxalase/ bleomycin resistance protein/ dioxygenase from Nocardioides.
To be Published
|
|
5BQF
 
 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L(+)-tartaric acid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obaidi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L-tartaric acid
to be published
|
|
4QS5
 
 | | CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE LIGW2 FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 (TARGET EFI-505250) WITH BOUND MANGANESE AND 3-methoxy-4-hydroxy-5-nitrobenzoic acid, THE D314N MUTANT | | Descriptor: | 4-hydroxy-3-methoxy-5-nitrobenzoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Raushel, M, Almo, S.C. | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of 5-CARBOXYVANILLATE Decarboxylase from Novosphingobium Aromaticivorans
To be Published
|
|
4NOF
 
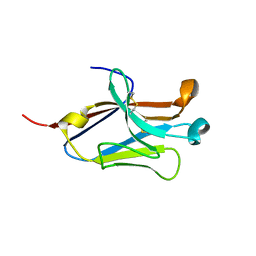 | | Crystal structure of the second Ig domain from mouse Polymeric Immunoglobulin receptor [PSI-NYSGRC-006220] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Polymeric immunoglobulin receptor | | Authors: | Sampathkumar, P, Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the second Ig domain from mouse Polymeric Immunoglobulin receptor
to be published
|
|
4Y9T
 
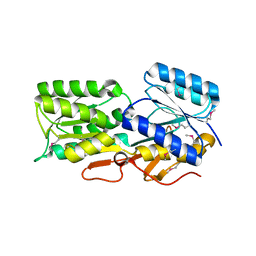 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GLUCOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, ABC transporter, solute binding protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-02-17 | | Release date: | 2015-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4N5U
 
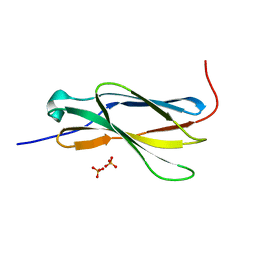 | | Crystal structure of the 4th FN3 domain of human Protein Tyrosine phosphatase, receptor type F [PSI-NYSGRC-006240] | | Descriptor: | Receptor-type tyrosine-protein phosphatase F, SULFATE ION | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.456 Å) | | Cite: | Crystal structure of the 4th FN3 domain of human PTP, receptor F [PSI-NYSGRC-006240]
to be published
|
|
5BR1
 
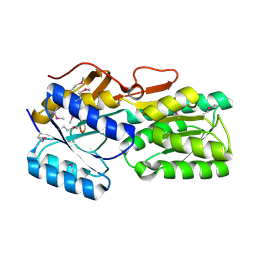 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GALACTOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-galactopyranose, ABC transporter, binding protein | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4NOB
 
 | | Crystal structure of the 1st Ig domain from mouse Polymeric Immunoglobulin receptor [PSI-NYSGRC-006220] | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of the 1st Ig domain from mouse Polymeric Immunoglobulin receptor
to be published
|
|
4E3Z
 
 | | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42 | | Descriptor: | Putative oxidoreductase protein | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-11 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
4DIO
 
 | | The crystal structure of transhydrogenase from Sinorhizobium meliloti | | Descriptor: | NAD(P) transhydrogenase subunit alpha part 1 | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-31 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of transhydrogenase from Sinorhizobium meliloti
To be Published
|
|
4HL9
 
 | | Crystal structure of antibiotic biosynthesis monooxygenase | | Descriptor: | Antibiotic biosynthesis monooxygenase | | Authors: | Rice, S, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-16 | | Release date: | 2012-10-31 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of antibiotic biosynthesis monooxygenase
To be Published
|
|
4HY3
 
 | | Crystal structure of a phosphoglycerate oxidoreductase from rhizobium etli | | Descriptor: | phosphoglycerate oxidoreductase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-11-12 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a phosphoglycerate oxidoreductase from rhizobium etli
To be Published
|
|
