5OGI
 
 | | Complex of a binding protein and human adenovirus C 5 hexon | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-07-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
1CPB
 
 | |
6EQC
 
 | | Cryo-EM reconstruction of a complex of a binding protein and human adenovirus C5 hexon | | Descriptor: | Hexon protein, scFv of 9C12 antibody | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-02-07 | | Last modified: | 2018-02-14 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
5AEF
 
 | | Electron cryo-microscopy of an Abeta(1-42)amyloid fibril | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Schmidt, M, Rohou, A, Lasker, K, Yadav, J.K, Schiene-Fischer, C, Fandrich, M, Grigorieff, N. | | Deposit date: | 2015-08-29 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Peptide Dimer Structure in an Abeta(1-42) Fibril Visualized with Cryo-Em
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6P73
 
 | | Cytochrome-C-nitrite reductase | | Descriptor: | CALCIUM ION, Cytochrome c-552, HEME C | | Authors: | Schmidt, M, Pacheco, A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Trapping of a Putative Intermediate in the CytochromecNitrite Reductase (ccNiR)-Catalyzed Reduction of Nitrite: Implications for the ccNiR Reaction Mechanism.
J.Am.Chem.Soc., 141, 2019
|
|
7JRI
 
 | |
7JR5
 
 | | Real Time Reaction Intermediates in Stigmatella Bacteriophytochrome P2 | | Descriptor: | 3-[2-[[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-5-[[(3~{S})-4-ethyl-3-methyl-2-oxidanylidene-1,3-dihydropyrrol-5-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, BENZAMIDINE, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M. | | Deposit date: | 2020-08-11 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structures of transient intermediates in the phytochrome photocycle.
Structure, 29, 2021
|
|
8ADE
 
 | |
2BW9
 
 | | Laue Structure of L29W MbCO | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schmidt, M, Nienhaus, K, Pahl, R, Krasselt, A, Anderson, S, Parak, F, Nienhaus, G.U, Srajer, V. | | Deposit date: | 2005-07-13 | | Release date: | 2005-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Ligand Migration Pathway and Protein Dynamics in Myoglobin: A Time-Resolved Crystallographic Study on L29W Mbco.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4K71
 
 | | Crystal structure of a high affinity Human Serum Albumin variant bound to the Neonatal Fc Receptor | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, SULFATE ION, ... | | Authors: | Schmidt, M.M, Townson, S.A, Andreucci, A, Dombrowski, C, Erbe, D.V, King, B, Kovalchin, J.T, Masci, A, Murillo, A, Schirmer, E.B, Furfine, E.S, Barnes, T.M. | | Deposit date: | 2013-04-16 | | Release date: | 2013-10-23 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an HSA/FcRn complex reveals recycling by competitive mimicry of HSA ligands at a pH-dependent hydrophobic interface.
Structure, 21, 2013
|
|
6BAK
 
 | | The structure of the Stigmatella aurantiaca phytochrome chromophore binding domain T289H mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-13 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
4CS3
 
 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in complex with an adenylated furan-bearing noncanonical amino acid and pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-(furan-2-yl)ethyl hydrogen carbonate, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
4CS4
 
 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in complex with AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, 2-{[dihydroxy(4-aminoethylphenyl)-{4}-sulfanyl]amino}-3-hydroxypropanoic acid, MAGNESIUM ION, ... | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
2BWH
 
 | | Laue Structure of a Short Lived State of L29W Myoglobin | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schmidt, M, Nienhaus, K, Pahl, R, Krasselt, A, Anderson, S, Parak, F, Nienhaus, G.U, Srajer, V. | | Deposit date: | 2005-07-14 | | Release date: | 2005-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand migration pathway and protein dynamics in myoglobin: a time-resolved crystallographic study on L29W MbCO.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
4CS2
 
 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in its apo form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PYRROLYSINE--TRNA LIGASE | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
6BAP
 
 | | Stigmatella aurantiaca bacterial phytochrome PAS-GAF-PHY, T289H mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAY
 
 | | Stigmatella aurantiaca bacterial phytochrome P1, PAS-GAF-PHY T289H mutant, room temperature structure | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-16 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAO
 
 | | Stigmatella aurantiaca phytochrome photosensory core module, wild type | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
2J96
 
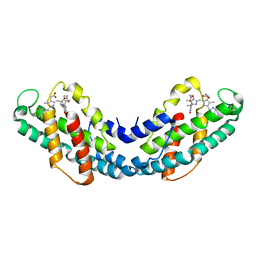 | | The E-configuration of alfa-Phycoerythrocyanin | | Descriptor: | PHYCOERYTHROCYANIN ALPHA CHAIN, PHYCOVIOLOBILIN | | Authors: | Schmidt, M, Patel, A, Zhao, Y, Reuter, W. | | Deposit date: | 2006-11-02 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for the Photochemistry of Alfa-Phycoerythrocyanin
Biochemistry, 46, 2007
|
|
7JVZ
 
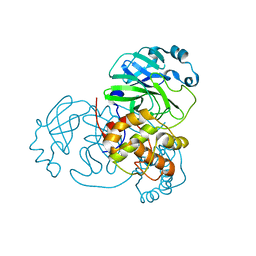 | |
6BAF
 
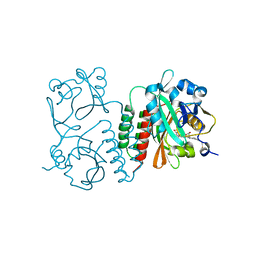 | | Structure of the chromophore binding domain of Stigmatella aurantiaca phytochrome P1, wild-type | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
1AR5
 
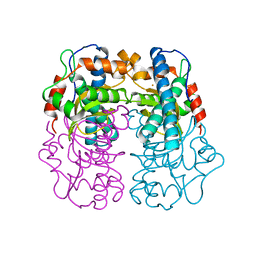 | |
1AR4
 
 | |
2C7K
 
 | | Laue structure of phycoerythrocyanin from Mastigocladus laminosus | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, PHYCOERYTHROCYANIN ALPHA CHAIN, ... | | Authors: | Schmidt, M, Krasselt, A, Reuter, W. | | Deposit date: | 2005-11-24 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Local Protein Flexibility as a Prerequisite for Reversible Chromophore Isomerization in Alpha-Phycoerythrocyanin
Biochim.Biophys.Acta, 1764, 2006
|
|
2C7J
 
 | | Phycoerythrocyanin from Mastigocladus laminosus, 295 K, 3.0 A | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, PHYCOERYTHROCYANIN ALPHA CHAIN, ... | | Authors: | Schmidt, M, Krasselt, A, Reuter, W. | | Deposit date: | 2005-11-24 | | Release date: | 2006-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Local Protein Flexibility as a Prerequisite for Reversible Chromophore Isomerization in Alfa- Phycoerythrocyanin
Biochim.Biophys.Acta, 1764, 2006
|
|
