6SFF
 
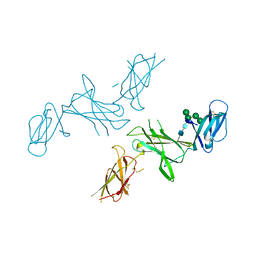 | | mouse Interleukin-12 subunit beta - p80 homodimer in space group I41 | | Descriptor: | CHLORIDE ION, Interleukin-12 subunit beta, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Homogeneously N-glycosylated proteins derived from the GlycoDelete HEK293 cell line enable diffraction-quality crystallogenesis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5D22
 
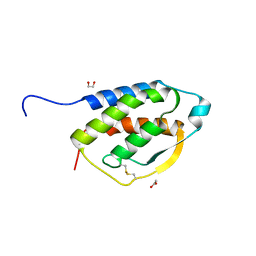 | |
7OPB
 
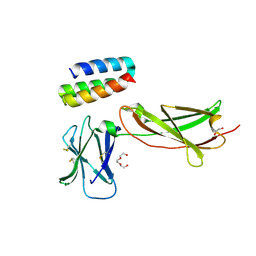 | | IL7R in complex with an antagonist | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IL7R binder, ... | | Authors: | Markovic, I, Verschueren, K.H.G, Verstraete, K, Savvides, S.N. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
3LN6
 
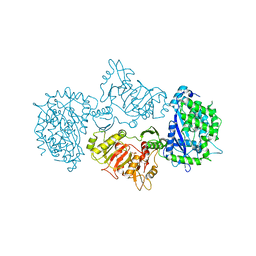 | |
1G6O
 
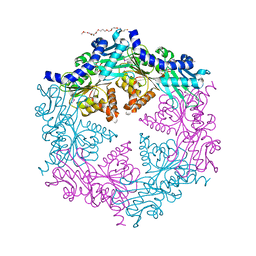 | | CRYSTAL STRUCTURE OF THE HELICOBACTER PYLORI ATPASE, HP0525, IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAG-ALPHA, DI(HYDROXYETHYL)ETHER | | Authors: | Yeo, H.J, Savvides, S.N, Herr, A.B, Lanka, E, Waksman, G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-11-07 | | Release date: | 2001-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system.
Mol.Cell, 6, 2000
|
|
5K1K
 
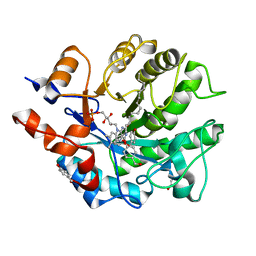 | | Crystal structure of oxidized Shewanella Yellow Enzyme 4 (SYE4) in complex with p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, NAD(P)H:flavin oxidoreductase Sye4, Octadecane, ... | | Authors: | Elegheert, J, Brige, A, Savvides, S.-N. | | Deposit date: | 2016-05-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Structural dissection of Shewanella oneidensis old yellow enzyme 4 bound to a Meisenheimer complex and (nitro)phenolic ligands.
FEBS Lett., 591, 2017
|
|
3M8U
 
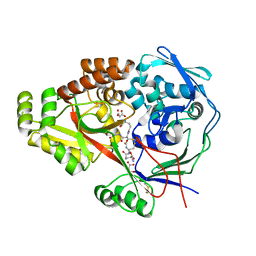 | | Crystal structure of glutathione-binding protein A (GbpA) from Haemophilus parasuis SH0165 in complex with glutathione disulfide (GSSG) | | Descriptor: | Heme-binding protein A, MALONATE ION, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Vergauwen, B, Elegheert, J, Dansercoer, A, Devreese, B, Savvides, S.N. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glutathione import in Haemophilus influenzae Rd is primed by the periplasmic heme-binding protein HbpA
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1DNC
 
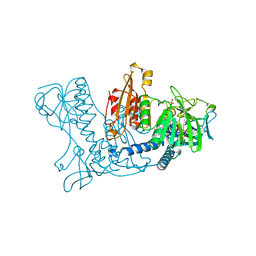 | | HUMAN GLUTATHIONE REDUCTASE MODIFIED BY DIGLUTATHIONE-DINITROSO-IRON | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GLUTATHIONE REDUCTASE, ... | | Authors: | Becker, K, Savvides, S.N, Keese, M, Schirmer, R.H, Karplus, P.A. | | Deposit date: | 1998-02-20 | | Release date: | 1998-05-27 | | Last modified: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers.
Nat.Struct.Biol., 5, 1998
|
|
2RAB
 
 | | Structure of glutathione amide reductase from Chromatium gracile in complex with NAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NICKEL (II) ION, ... | | Authors: | Van Petegem, F, De Vos, D, Savvides, S, Vergauwen, B, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
6ZL1
 
 | | Crystal structure of human serum albumin in complex with the MCL-1 neutralizing Alphabody CMPX-383B | | Descriptor: | Albumin, CMPX-383B | | Authors: | Pannecoucke, E, Savvides, S.N, Desmet, J, Lasters, I. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.272 Å) | | Cite: | Cell-penetrating Alphabody protein scaffolds for intracellular drug targeting.
Sci Adv, 7, 2021
|
|
6ZIE
 
 | | Crystal structure of MCL-1 in complex with a neutralizing Alphabody CMPX-383B | | Descriptor: | CMPX-383B, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Pannecoucke, E, Savvides, S.N, Desmet, J, Lasters, I. | | Deposit date: | 2020-06-25 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cell-penetrating Alphabody protein scaffolds for intracellular drug targeting.
Sci Adv, 7, 2021
|
|
5J12
 
 | |
6SMC
 
 | | mouse Interleukin-12 subunit beta - p80 homodimer in space group P1 | | Descriptor: | CHLORIDE ION, Interleukin-12 subunit beta, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2019-08-21 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Around she goes: the structure of mouse Interleukin-12 p80
To Be Published
|
|
7OX5
 
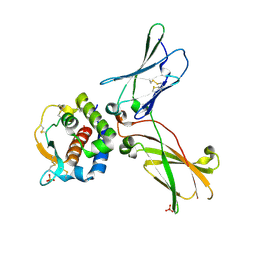 | | hIL-9:hIL-9Ra complex | | Descriptor: | Interleukin-9, Interleukin-9 receptor, SULFATE ION | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
6SP3
 
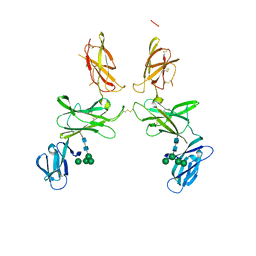 | | mouse Interleukin-12 subunit beta - p80 homodimer in space group P21 crystal form 1 | | Descriptor: | CHLORIDE ION, Interleukin-12 subunit beta, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Around she goes: the structure of mouse Interleukin-12 p80
To Be Published
|
|
7AL7
 
 | | The Crystal Structure of Human IL-18 in Complex With Human IL-18 Binding Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutathione S-transferase class-mu 26 kDa isozyme,Interleukin-18, Interleukin-18-binding protein | | Authors: | Detry, S, Andries, J, Bloch, Y, Clancy, D, Savvides, S.N. | | Deposit date: | 2020-10-05 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of human IL-18 sequestration by the decoy receptor IL-18 binding protein in inflammation and tumor immunity.
J.Biol.Chem., 298, 2022
|
|
5J13
 
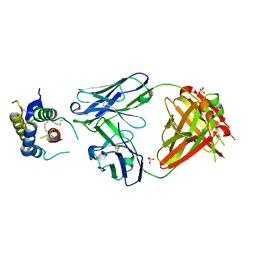 | |
5J11
 
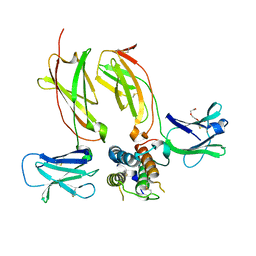 | | Structure of human TSLP in complex with TSLPR and IL-7Ralpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor-like factor 2, Interleukin-7 receptor subunit alpha, ... | | Authors: | Verstraete, K, Savvides, S.N. | | Deposit date: | 2016-03-28 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure and antagonism of the receptor complex mediated by human TSLP in allergy and asthma.
Nat Commun, 8, 2017
|
|
7OX2
 
 | | Fab 6E2: hIL-9 complex | | Descriptor: | Heavy chain (Fab 6E2), Interleukin-9, Light chain (Fab 6E2), ... | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
2R9Z
 
 | | Glutathione amide reductase from Chromatium gracile | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione amide reductase, ... | | Authors: | Van Petegem, F, Vergauwen, B, Savvides, S, De Vos, D, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
7PQH
 
 | | Cryo-EM structure of Saccharomyces cerevisiae TOROID (TORC1 Organized in Inhibited Domains). | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 1 subunit KOG1,Target of rapamycin complex 1 subunit Kog1, Target of rapamycin complex subunit LST8 | | Authors: | Felix, J, Prouteau, M, Bourgoint, C, Bonadei, L, Desfosses, A, Gabus, C, Sadian, Y, Savvides, S.N, Gutsche, I, Loewith, R. | | Deposit date: | 2021-09-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | EGOC inhibits TOROID polymerization by structurally activating TORC1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7OX4
 
 | | Mouse interleukin-9 in complex with Fab 35D8. | | Descriptor: | ACETATE ION, Heavy chain (Fab 35D8), Interleukin-9, ... | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
7OX1
 
 | | Fab 7D6: hIL-9 complex | | Descriptor: | Heavy chain (Fab 7D6), Interleukin-9, Light chain (Fab 7D6) | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
7ZV9
 
 | |
8AD2
 
 | | Tobacco lectin Nictaba in complex with triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Nictaba, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2022-07-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the archetypical Nictaba plant lectin reveals the molecular basis for its carbohydrate-binding properties
Biorxiv, 2024
|
|
