1QW2
 
 | | Crystal Structure of a Protein of Unknown Function TA1206 from Thermoplasma acidophilum | | Descriptor: | conserved hypothetical protein TA1206 | | Authors: | Savchenko, A, Evdokimova, E, Kudrytska, M, Edwards, A.E, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-30 | | Release date: | 2004-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Hypothetical Protein "TA1206" from Thermoplasma acidophilum
To be Published
|
|
1KR4
 
 | | Structure Genomics, Protein TM1056, cutA | | Descriptor: | Protein TM1056, cutA | | Authors: | Savchenko, A, Zhang, R, Joachimiak, A, Edwards, A, Akarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of CutA from Thermotoga maritima at 1.4 A resolution.
Proteins, 54, 2004
|
|
1P9Q
 
 | | Structure of a hypothetical protein AF0491 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0491 | | Authors: | Savchenko, A, Evdokimova, E, Skarina, T, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
7MH7
 
 | |
7M92
 
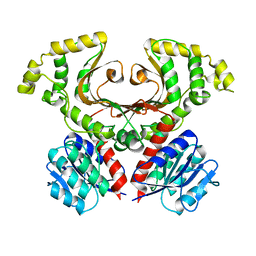 | |
6OX6
 
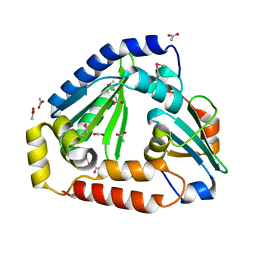 | | Crystal structure of the complex between the Type VI effector Tas1 and its immunity protein | | Descriptor: | ACETATE ION, PA14_01140, Tas1 | | Authors: | Ahmad, S, Stogios, P.J, Skarina, T, Whitney, J, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp.
Nature, 575, 2019
|
|
6OZ1
 
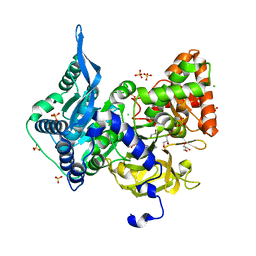 | | Crystal structure of the adenylation (A) domain of the carboxylate reductase (CAR) GR01_22995 from Mycobacterium chelonae | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Fedorchuk, T, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | One-Pot Biocatalytic Transformation of Adipic Acid to 6-Aminocaproic Acid and 1,6-Hexamethylenediamine Using Carboxylic Acid Reductases and Transaminases.
J.Am.Chem.Soc., 142, 2020
|
|
2KKY
 
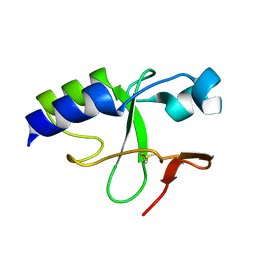 | | Solution Structure of C-terminal domain of oxidized NleG2-3 (residue 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | Descriptor: | Uncharacterized protein ECs2156 | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
9MTO
 
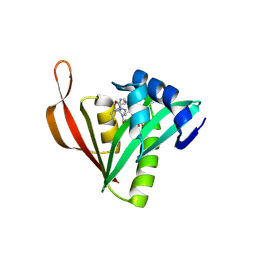 | | Crystal structure of acetyltransferase CBU0801 from Coxiella burnetii | | Descriptor: | COENZYME A, [Ribosomal protein bS18]-alanine N-acetyltransferase | | Authors: | Stogios, P.J, Evdokimova, E, Chang, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2025-01-11 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of acetyltransferase CBU0801 from Coxiella burnetii
To Be Published
|
|
4W8K
 
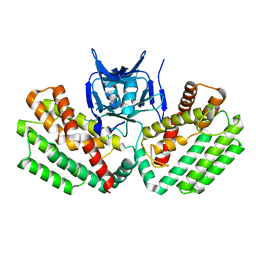 | | Crystal structure of a putative Cas1 enzyme from Vibrio phage ICP1 | | Descriptor: | Cas1 protein, POTASSIUM ION | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyeno, O, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To Be Published
|
|
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | Descriptor: | Probable sphingosine-1-phosphate lyase | | Authors: | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4W97
 
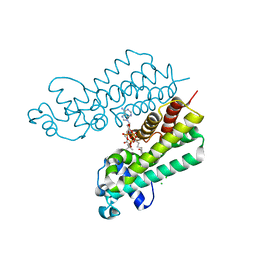 | | Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, HTH-type transcriptional repressor KstR2, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 3-[(3aS,4S,7aS)-7a-methyl-1,5-bis(oxidanylidene)-2,3,3a,4,6,7-hexahydroinden-4-yl]propanethioate | | Authors: | Stogios, P.J, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Ketosteroid Transcriptional Regulator of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
6UXT
 
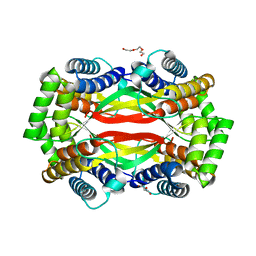 | | Crystal structure of unknown function protein yfdX from Shigella flexneri | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Chang, C, Skarina, T, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-08 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Crystal structure of unknown function protein yfdX from Shigella flexneri
To Be Published
|
|
6UVZ
 
 | | Amidohydrolase 2 from Bifidobacterium longum subsp. infantis | | Descriptor: | Amidohydrolase 2, CITRIC ACID, NONAETHYLENE GLYCOL | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-11-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Amidohydrolase 2 from Bifidobacterium longum subsp. infantis
To Be Published
|
|
1PW5
 
 | | putative nagD protein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, SULFATE ION, nagD protein, ... | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-06-30 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | putative nagD protein
To be Published
|
|
8SCD
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with reaction intermediate | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, 4-AMINOBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
8SDC
 
 | | Crystal structure of fluoroacetate dehalogenase Daro3835 apoenzyme | | Descriptor: | Alpha/beta hydrolase fold protein, CHLORIDE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutdinova, A, Iakounine, A, Savchenko, A. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into hydrolytic defluorination of difluoroacetate by microbial fluoroacetate dehalogenases.
Febs J., 290, 2023
|
|
6UX3
 
 | | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae | | Descriptor: | Acetoin dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Chang, C, Skarina, T, Mesa, N, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-06 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae
To Be Published
|
|
1Q7H
 
 | | Structure of a Conserved PUA Domain Protein from Thermoplasma acidophilum | | Descriptor: | ZINC ION, conserved hypothetical protein | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-18 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a conserved hypothetical protein from T. acidophilum
TO BE PUBLISHED
|
|
8VVA
 
 | |
6HCD
 
 | | Structure of universal stress protein from Archaeoglobus fulgidus | | Descriptor: | ACETATE ION, CHLORIDE ION, UNIVERSAL STRESS PROTEIN, ... | | Authors: | Shumilin, I.A, Loch, J.I, Cymborowski, M, Xu, X, Edwards, A, Di Leo, R, Shabalin, I.G, Joachimiak, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-08-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
4RXV
 
 | | The crystal structure of the N-terminal fragment of uncharacterized protein from Legionella pneumophila | | Descriptor: | hypothetical protein lpg0944 | | Authors: | Nocek, B, Cuff, M, Evdokimova, E, Egorova, O, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-12 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
4U12
 
 | | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer | | Descriptor: | Uncharacterized protein HP0242 | | Authors: | Grabowski, M, Shabalin, I.G, Chruszcz, M, Skarina, T, Onopriyenko, O, Guthrie, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-14 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer
to be published
|
|
8VS5
 
 | | Structure of catalytic domain of telomere resolvase, ResT, from Borrelia garinii | | Descriptor: | Telomere resolvase ResT | | Authors: | Semper, C, Savchenko, A, Watanabe, N, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure analysis of the telomere resolvase from the Lyme disease spirochete Borrelia garinii reveals functional divergence of its C-terminal domain.
Nucleic Acids Res., 52, 2024
|
|
8VJ1
 
 | | Structure of C-terminal domain of telomere resolvase, ResT, from Borrelia garinii | | Descriptor: | Telomere resolvase ResT | | Authors: | Semper, C, Savchenko, A, Watanabe, N, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure analysis of the telomere resolvase from the Lyme disease spirochete Borrelia garinii reveals functional divergence of its C-terminal domain.
Nucleic Acids Res., 52, 2024
|
|
