8UU6
 
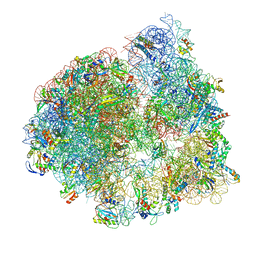 | | Cryo-EM structure of the ratcheted Listeria innocua 70S ribosome in complex with p/E-tRNA (structure II-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UUA
 
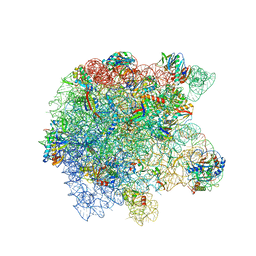 | | Cryo-EM structure of the Listeria innocua 50S ribosomal subunit in complex with HflXr (structure III) | | Descriptor: | 23S Ribosomal RNA, 5S Ribosomal RNA, GTPase HflX, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU5
 
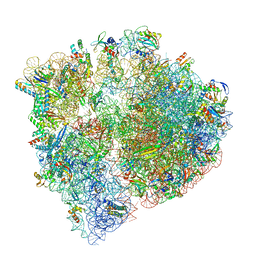 | | Cryo-EM structure of the Listeria innocua 70S ribosome (head-swiveled) in complex with pe/E-tRNA (structure I-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU9
 
 | | Cryo-EM structure of the ratcheted Listeria innocua 70S ribosome (head-swiveled) in complex with HflXr and pe/E-tRNA (structure II-D) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU7
 
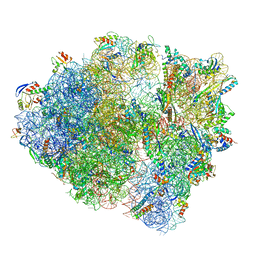 | | Cryo-EM structure of the Listeria innocua 70S ribosome in complex with HflXr, HPF, and E-site tRNA (structure II-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU8
 
 | | Cryo-EM structure of the Listeria innocua 70S ribosome (head-swiveled) in complex with HflXr and pe/E-tRNA (structure II-C) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UHG
 
 | |
8UI0
 
 | |
8UIS
 
 | | Structure of transcription complex Pol II-DSIF-NELF-TFIIS | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Su, B.G, Vos, S.M. | | Deposit date: | 2023-10-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing.
Mol.Cell, 84, 2024
|
|
8UHA
 
 | |
8UHD
 
 | | Structure of paused transcription complex Pol II-DSIF-NELF - post-translocated | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Su, B.G, Vos, S.M. | | Deposit date: | 2023-10-08 | | Release date: | 2024-03-20 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing.
Mol.Cell, 84, 2024
|
|
8YT4
 
 | | Structure of Aquifex aeolicus Lumazine Synthase by Cryo-Electron Microscopy to 1.42 Angstrom Resolution | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Savva, C.G, Sobhy, M.A, De Biasio, A, Hamdan, S.M. | | Deposit date: | 2024-03-24 | | Release date: | 2024-04-10 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (1.42 Å) | | Cite: | Structure of Aquifex aeolicus lumazine synthase by cryo-electron microscopy to 1.42 angstrom resolution.
Iucrj, 11, 2024
|
|
8VXQ
 
 | |
8UU4
 
 | | Cryo-EM structure of the Listeria innocua 70S ribosome in complex with HPF (structure I-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8VT0
 
 | | SPOT-RASTR - a cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Esfahani, B.G, Randolph, P, Peng, R, Grant, T, Stroupe, M.E, Stagg, S.M. | | Deposit date: | 2024-01-25 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | SPOT-RASTR-A cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface.
Pnas Nexus, 3, 2024
|
|
8XPB
 
 | | Crystal structure of d(ACGCCGT/ACGGCGT) in complex with Echinomycin | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*CP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3'), ... | | Authors: | Hou, M.H, Huang, H.T, Lin, S.M, Neidle, S. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
8XWX
 
 | |
8XP8
 
 | | Crystal structure of d(ACGmCCGT/ACGGCGT) in complex with Echinomycin | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*(5CM)P*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3'), ... | | Authors: | Hou, M.H, Lin, S.M, Neidle, H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
8XP9
 
 | | Crystal structure of d(ACGCCGT/ACGGCGT) | | Descriptor: | DNA (5'-D(P*AP*CP*GP*CP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3') | | Authors: | Hou, M.H, Lin, S.M, Lin, Y.J, Neidle, S. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
8XPA
 
 | | Crystal structure of d(ACGmCCGT/ACGGCGT) | | Descriptor: | DNA (5'-D(P*AP*CP*GP*(5CM)P*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Hou, M.H, Lin, S.M, Neidle, S. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
4RIS
 
 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain, Envelope glycoprotein | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
4RIR
 
 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
1CJB
 
 | | MALARIAL PURINE PHOSPHORIBOSYLTRANSFERASE | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, MAGNESIUM ION, PROTEIN (HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE), ... | | Authors: | Shi, W, Li, C.M, Tyler, P.C, Furneaux, R.H, Cahill, S.M, Girvin, M.E, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-04-08 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of malarial purine phosphoribosyltransferase in complex with a transition-state analogue inhibitor.
Biochemistry, 38, 1999
|
|
8GCG
 
 | | MDM2 bound to inhibitor | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, macrocyclic peptide inhibitor | | Authors: | Silvestri, A.P, Muir, E.W, Chakka, S.K, Tripathi, S.M, Rubin, S.M, Pye, C.R, Schwochert, J.A. | | Deposit date: | 2023-03-01 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | DNA-Encoded Macrocyclic Peptide Libraries Enable the Discovery of a Neutral MDM2-p53 Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
6P8F
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
