2PCP
 
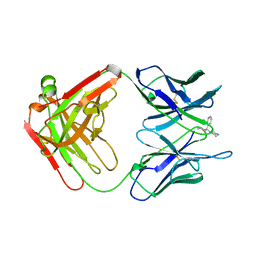 | | ANTIBODY FAB COMPLEXED WITH PHENCYCLIDINE | | Descriptor: | 1-(PHENYL-1-CYCLOHEXYL)PIPERIDINE, IMMUNOGLOBULIN | | Authors: | Lim, K, Owens, S.M, Arnold, L, Sacchettini, J.C, Linthicum, D.S. | | Deposit date: | 1998-06-02 | | Release date: | 1999-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of monoclonal 6B5 Fab complexed with phencyclidine.
J.Biol.Chem., 273, 1998
|
|
2PDE
 
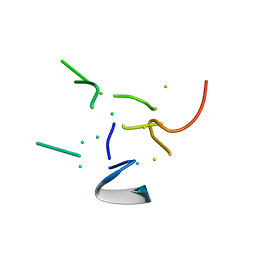 | | THE HIGH RESOLUTION STRUCTURE OF THE PERIPHERAL SUBUNIT-BINDING DOMAIN OF DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM THE PYRUVATE DEHYDROGENASE MULTIENZYME COMPLEX OF BACILLUS STEAROTHERMOPHILUS | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | Authors: | Kalia, Y.N, Brocklehurst, S.M, Hipps, D.S, Appella, E, Sakaguchi, K, Perham, R.N. | | Deposit date: | 1992-11-25 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The high-resolution structure of the peripheral subunit-binding domain of dihydrolipoamide acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus.
J.Mol.Biol., 230, 1993
|
|
6XKR
 
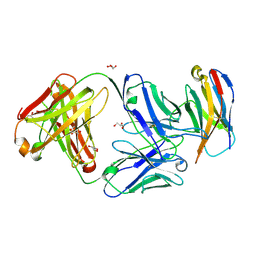 | | Structure of Sasanlimab Fab in complex with PD-1 | | Descriptor: | GLYCEROL, Programmed cell death protein 1, Sasanlimab Fab Heavy chain, ... | | Authors: | Kimberlin, C.R, Chin, S.M. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Pharmacologic Properties and Preclinical Activity of Sasanlimab, A High-affinity Engineered Anti-Human PD-1 Antibody.
Mol.Cancer Ther., 19, 2020
|
|
6ETI
 
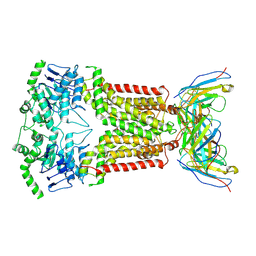 | | Structure of inhibitor-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Altmann, K.H, Locher, K.P. | | Deposit date: | 2017-10-26 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5NJG
 
 | | Structure of an ABC transporter: part of the structure that could be built de novo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | Authors: | Taylor, N.M.I, Manolaridis, I, Jackson, S.M, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure of the human multidrug transporter ABCG2.
Nature, 546, 2017
|
|
6RRV
 
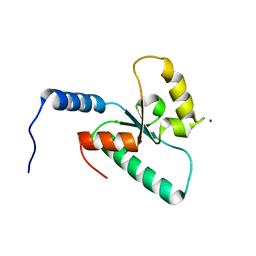 | | Crystal structure of the Sir4 H-BRCT domain | | Descriptor: | BROMIDE ION, CHLORIDE ION, Regulatory protein SIR4 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
7T8W
 
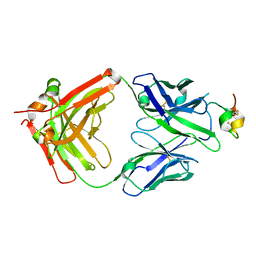 | | Structure of antibody 3G12 bound to Respiratory Syncytial Virus G central conserved domain mutant S177Q | | Descriptor: | 3G12 Fab Heavy chain, 3G12 Fab Light chain, Mature secreted glycoprotein G | | Authors: | Nunez Castrejon, A.M, O'Rourke, S.M, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Design and Antigenic Validation of Respiratory Syncytial Virus G Immunogens.
J.Virol., 96, 2022
|
|
1OJY
 
 | | Decay accelerating factor (cd55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OK2
 
 | | Decay accelerating factor (CD55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OJV
 
 | | Decay accelerating factor (CD55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, C.M, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7T7S
 
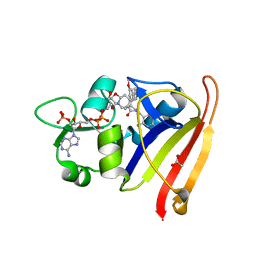 | | R-27 in Complex with S. aureus DHFR and tricyclic-NADPH (tNADPH) | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, ACETATE ION, Dihydrofolate reductase, ... | | Authors: | Reeve, S.M, Wang, S, Donald, B.R, Wright, D.L. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chiral evasion and stereospecific antifolate resistance in Staphylococcus aureus.
Plos Comput.Biol., 18, 2022
|
|
1OK1
 
 | | Decay accelerating factor (cd55) : the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2PF0
 
 | |
6RUU
 
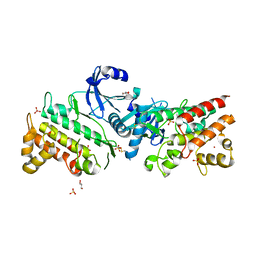 | | Pseudokinase domain of human IRAK3 | | Descriptor: | GLYCEROL, Interleukin-1 receptor-associated kinase 3, MERCURY (II) ION, ... | | Authors: | Lange, S.M, Kulathu, Y, Cohen, P. | | Deposit date: | 2019-05-29 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dimeric Structure of the Pseudokinase IRAK3 Suggests an Allosteric Mechanism for Negative Regulation.
Structure, 29, 2021
|
|
1OK3
 
 | | Decay accelerating factor (cd55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7T7Q
 
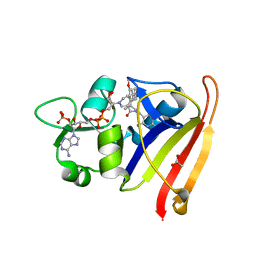 | | R-27 In Complex with S. aureus DHFR and alpha-NADPH - Remediated for comparison with tNADPH | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, ACETATE ION, Dihydrofolate reductase, ... | | Authors: | Reeve, S.M, Wang, S, Donald, B.R, Wright, D.L. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chiral evasion and stereospecific antifolate resistance in Staphylococcus aureus.
Plos Comput.Biol., 18, 2022
|
|
2OYS
 
 | | Crystal Structure of SP1951 protein from Streptococcus pneumoniae in complex with FMN, Northeast Structural Genomics Target SpR27 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Hypothetical protein SP1951 | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Janjua, H, Satterwhite, R, Liu, J, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
6F2D
 
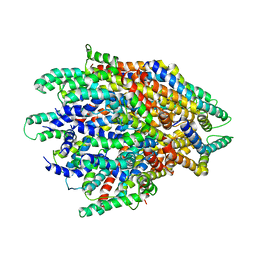 | | A FliPQR complex forms the core of the Salmonella type III secretion system export apparatus. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Johnson, S, Kuhlen, L, Abrusci, P, Lea, S.M. | | Deposit date: | 2017-11-24 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the core of the type III secretion system export apparatus.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2P28
 
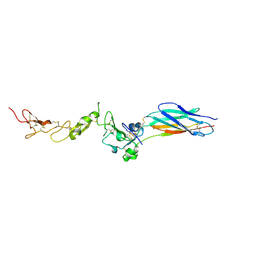 | | Structure of the PHE2 and PHE3 fragments of the integrin beta2 subunit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin beta-2 | | Authors: | Shi, M, Foo, S.Y, Tan, S.M, Mitchell, E.P, Law, S.K.A, Lescar, J. | | Deposit date: | 2007-03-07 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural hypothesis for the transition between bent and extended conformations of the leukocyte beta2 integrins
J.Biol.Chem., 282, 2007
|
|
6S1N
 
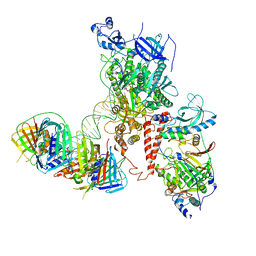 | | Human polymerase delta holoenzyme Conformer 2 | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-06-19 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
7T6C
 
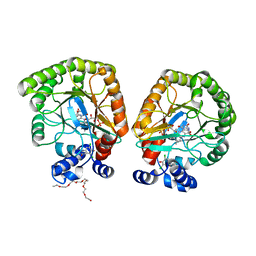 | | E. coli dihydroorotate dehydrogenase bound to the ubiquinone surrogate DCIP | | Descriptor: | 2,6-bis(chloranyl)-4-[(4-hydroxyphenyl)amino]phenol, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Horwitz, S.M, Ambarian, J.A, Davis, K.M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines.
Rsc Chem Biol, 3, 2022
|
|
6XF2
 
 | |
7T5K
 
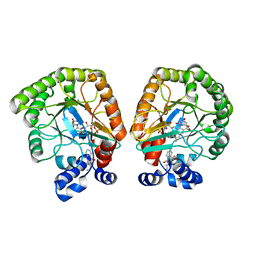 | | E. coli dihydroorotate dehydrogenase bound to the inhibitor HQNO | | Descriptor: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Horwitz, S.M, Ambarian, J.A, Davis, K.M. | | Deposit date: | 2021-12-12 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines.
Rsc Chem Biol, 3, 2022
|
|
7T6H
 
 | | E. coli dihydroorotate dehydrogenase | | Descriptor: | Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, OROTIC ACID | | Authors: | Horwitz, S.M, Ambarian, J.A, Davis, K.M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines.
Rsc Chem Biol, 3, 2022
|
|
7T5Y
 
 | | E. coli dihydroorotate dehydrogenase bound to the inhibitor HMNQ | | Descriptor: | 3-methyl-2-[(2E)-non-2-en-1-yl]quinolin-4(1H)-one, Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Horwitz, S.M, Ambarian, J.A, Davis, K.M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines.
Rsc Chem Biol, 3, 2022
|
|
