5DM2
 
 | |
5DPL
 
 | | The structure of PKMT2 from Rickettsia typhi in complex with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein lysine methyltransferase 2 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
7TPJ
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, Putative cell surface polysaccharide polymerase/ligase | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
7TPG
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, GERANYL DIPHOSPHATE, ... | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
7U36
 
 | | Crystal structure of human GSK3B in complex with ARN1484 | | Descriptor: | (3S)-1-[(2-fluorophenoxy)acetyl]-N-(pyridin-2-yl)pyrrolidine-3-carboxamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
5DA3
 
 | | Crystal structure of PTK6 Kinase domain with inhibitor | | Descriptor: | (2-chloro-4-{[6-cyclopropyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}phenyl)(morpholin-4-yl)methanone, GLYCEROL, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Battula, S.K, Vadivelu, S, Tyagi, R, Gosu, R. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structures of PTK6: With Dasatinib at 2.24 angstrom , with novel imidazo[1,2-a]pyrazin-8-amine derivative inhibitor at 1.70 angstrom resolution
Biochem. Biophys. Res. Commun., 482, 2017
|
|
7U2Z
 
 | | Crystal structure of human GSK3B in complex with G12 | | Descriptor: | (3R)-1-[3-(2-fluorophenyl)propanoyl]-N-(pyridin-2-yl)pyrrolidine-3-carboxamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
7U31
 
 | | Crystal structure of human GSK3B in complex with G5 | | Descriptor: | 5-(4-fluorophenyl)-4-[1-(methanesulfonyl)azetidin-3-yl]pyrimidin-2-amine, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
7U33
 
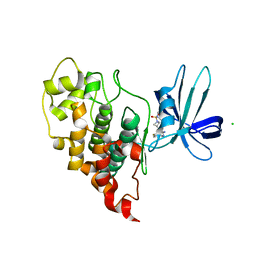 | | Crystal structure of human GSK3B in complex with ARN9133 | | Descriptor: | 3-[2-amino-5-(4-fluorophenyl)pyrimidin-4-yl]-N,N-dimethylazetidine-1-sulfonamide, CHLORIDE ION, Glycogen synthase kinase-3 beta | | Authors: | Tripathi, S.K, Balboni, B, Giabbai, B, Storici, P, Girotto, S, Cavalli, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Novel GSK-3 beta Hits Using Competitive Biophysical Assays.
Int J Mol Sci, 23, 2022
|
|
8CZK
 
 | |
8D19
 
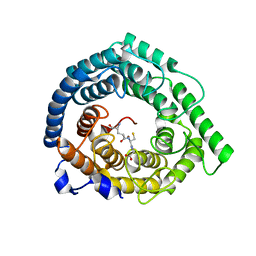 | | Human LanCL1 bound to GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D0V
 
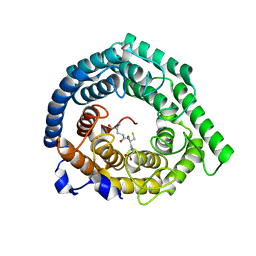 | | Human LanCL1 C264A mutant bound to GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8CZL
 
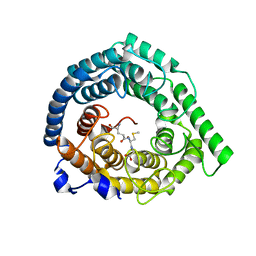 | | Human LanCL1 bound to methyl glutathione (MeGSH) | | Descriptor: | Glutathione S-transferase LANCL1, L-GAMMA-GLUTAMYL-S-METHYLCYSTEINYLGLYCINE, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8CUY
 
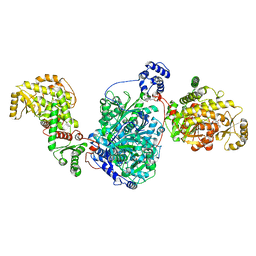 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CV0
 
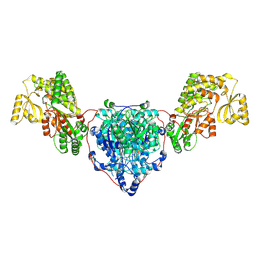 | | KS-AT domains of mycobacterial Pks13 with outward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CV1
 
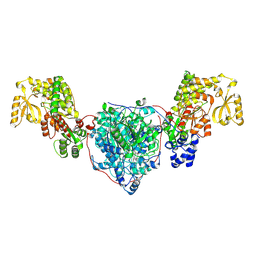 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CUZ
 
 | | KS-AT domains of mycobacterial Pks13 with inward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1YDB
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
5DM4
 
 | | Crystal structure of the plantazolicin methyltransferase BpumL in complex with pentazolic desmethylPZN analog and SAH | | Descriptor: | 1-[(4S)-4-(4-{4-[4-(5,5'-dimethyl-2,4'-bi-1,3-oxazol-2'-yl)-1,3-thiazol-2-yl]-5-methyl-1,3-oxazol-2-yl}-1,3-thiazol-2-yl)-4-(methylamino)butyl]guanidine, GLYCEROL, Methyltransferase domain family, ... | | Authors: | Hao, Y, Nair, S.K. | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into methyltransferase specificity and bioactivity of derivatives of the antibiotic plantazolicin.
Acs Chem.Biol., 10, 2015
|
|
5DPD
 
 | | The structure of PKMT1 from Rickettsia prowazekii in complex with AdoMet | | Descriptor: | S-ADENOSYLMETHIONINE, protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
1YDA
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
2GGE
 
 | | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, yitF | | Authors: | Malashkevich, V.N, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A
To be Published
|
|
2GHE
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, cytosolic ascorbate peroxidase 1 | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
1XU2
 
 | | The crystal structure of APRIL bound to BCMA | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Hymowitz, S.G, Patel, D.R, Wallweber, H.J.A, Runyon, S, Yan, M, Yin, J, Shriver, S.K, Gordon, N.C, Pan, B, Skelton, N.J, Kelley, R.F, Starovasnik, M.A. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of APRIL-receptor complexes: Like BCMA, TACI employs only a single cysteine-rich domain for high-affinity ligand binding
J.Biol.Chem., 280, 2005
|
|
1YW2
 
 | | Mutated Mus Musculus P38 Kinase (mP38) | | Descriptor: | 2-(ETHOXYMETHYL)-4-(4-FLUOROPHENYL)-3-[2-(2-HYDROXYPHENOXY)PYRIMIDIN-4-YL]ISOXAZOL-5(2H)-ONE, Mitogen-activated protein kinase 14 | | Authors: | Laughlin, S.K, Clark, M.P, Djung, J.F, Golebiowski, A, Brugel, T.A, Sabat, M, Bookland, R.G, Laufersweiler, M.J, Vanrens, J.C, Townes, J.A, De, B, Hsieh, L.C, Xu, S.C, Walter, R.L, Mekel, M.J, Janusz, M.J. | | Deposit date: | 2005-02-16 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The development of new isoxazolone based inhibitors of tumor necrosis factor-alpha (TNF-alpha) production.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
