7RC4
 
 | | Aeronamide N-methyltransferase, AerE (D141A) | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, Methyltransferase family protein, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC6
 
 | | Aeronamide N-methyltransferase, AerE, bound to modified peptide substrate, AerA-DL,34 | | Descriptor: | Aeronamide A peptide, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC5
 
 | | Aeronamide N-methyltransferase, AerE (N231A) | | Descriptor: | CALCIUM ION, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3K2G
 
 | | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, MAGNESIUM ION, Resiniferatoxin-binding, ... | | Authors: | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides
To be Published
|
|
5JGJ
 
 | | Crystal structure of GtmA | | Descriptor: | UbiE/COQ5 family methyltransferase, putative | | Authors: | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Doyle, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
5JI7
 
 | |
1WS4
 
 | | Crystal structure of Jacalin- Me-alpha-Mannose complex: Promiscuity vs Specificity | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, methyl alpha-D-galactopyranoside, ... | | Authors: | Jeyaprakash, A.A, Jayashree, G, Mahanta, S.K, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2004-10-31 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the energetics of jacalin-sugar interactions: promiscuity versus specificity
J.Mol.Biol., 347, 2005
|
|
5JI9
 
 | |
5JIU
 
 | |
7RI3
 
 | | Crystal structure of Albireti Toxin, a diphtheria toxin homolog, from Streptomyces albireticuli | | Descriptor: | ACETATE ION, CALCIUM ION, Diphtheria_T domain-containing protein, ... | | Authors: | Sugiman-Marangos, S.N, Gill, S.K, Melnyk, R.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of distant diphtheria toxin homologs reveal functional determinants of an evolutionarily conserved toxin scaffold.
Commun Biol, 5, 2022
|
|
5JIA
 
 | |
2F2K
 
 | | Aldose reductase tertiary complex with NADPH and DEG | | Descriptor: | Aldose reductase, GAMMA-GLUTAMYL-S-(1,2-DICARBOXYETHYL)CYSTEINYLGLYCINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Singh, R, White, M.A, Ramana, K.V, Petrash, J.M, Watowich, S.J, Bhatnagar, A, Srivastava, S.K. | | Deposit date: | 2005-11-17 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a glutathione conjugate bound to the active site of aldose reductase.
Proteins, 64, 2006
|
|
7RB4
 
 | | Crystal structure of Peptono Toxin, a diphtheria toxin homolog, from Seinonella peptonophila | | Descriptor: | Diphtheria toxin, C domain, IODIDE ION | | Authors: | Sugiman-Marangos, S.N, Gill, S.K, Melnyk, R.A. | | Deposit date: | 2021-07-05 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of distant diphtheria toxin homologs reveal functional determinants of an evolutionarily conserved toxin scaffold.
Commun Biol, 5, 2022
|
|
3KD9
 
 | |
2GGN
 
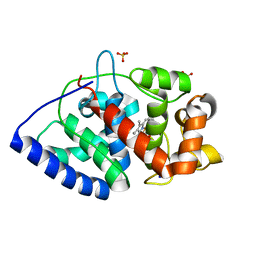 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, SULFATE ION, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
5IXM
 
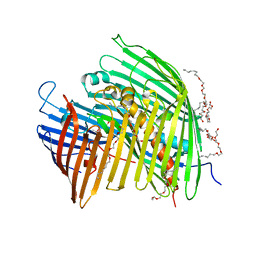 | | The LPS Transporter LptDE from Yersinia pestis, core complex | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD | | Authors: | Botos, I, Mayclin, S.J, McCarthy, J.G, Buchanan, S.K. | | Deposit date: | 2016-03-23 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens.
Structure, 24, 2016
|
|
3KCN
 
 | |
2GHD
 
 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
3KEW
 
 | | Crystal structure of probable alanyl-trna-synthase from Clostridium perfringens | | Descriptor: | DHHA1 domain protein, ZINC ION | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alanyl-trna-synthase from Clostridium perfringens
To be Published
|
|
3KFW
 
 | |
2G5U
 
 | | Human Transthyretin (TTR) Complexed with Hydroxylated polychlorinated Biphenyl-4,4'-dihydroxy-3,3',5,5'-tetrachlorobiphenyl | | Descriptor: | 3,5,3',5'-TETRACHLORO-BIPHENYL-4,4'-DIOL, Transthyretin | | Authors: | Palaninathan, S.K, Smith, C, Safe, S.H, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2006-02-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydroxylated polychlorinated biphenyls selectively bind transthyretin in blood and inhibit amyloidogenesis: rationalizing rodent PCB toxicity
Chem.Biol., 11, 2004
|
|
7RGG
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor BPTES | | Descriptor: | Glutaminase kidney isoform, mitochondrial 68 kDa chain, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
7REN
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor UPGL-00004 | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
2GDQ
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution | | Descriptor: | yitF | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-16 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Bacillus subtilis at 1.8 A resolution
To be Published
|
|
