4WY7
 
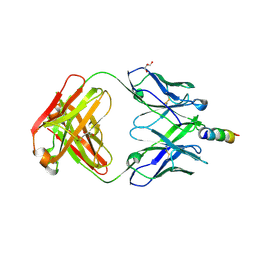 | | Crystal structure of recombinant 4E10 expressed in Escherichia coli with epitope bound | | Descriptor: | Envelope glycoprotein gp160, Fab 4E10 Heavy chain, Fab 4E10Light chain, ... | | Authors: | Rujas, E, Morante, K, Tsumoto, K, Nieva, J.L, Caaveiro, J.M.M. | | Deposit date: | 2014-11-16 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Atomic Structure of the HIV-1 gp41 Transmembrane Domain and Its Connection to the Immunogenic Membrane-proximal External Region.
J.Biol.Chem., 290, 2015
|
|
7JOO
 
 | | Crystal structure of ICOS in complex with antibody STIM003 and anti-kappa VHH domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Inducible T-cell costimulator, ... | | Authors: | Rujas, E, Sicard, T, Julien, J.P. | | Deposit date: | 2020-08-06 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of the ICOS/ICOS-L immune complex reveals high molecular mimicry by therapeutic antibodies.
Nat Commun, 11, 2020
|
|
6X4T
 
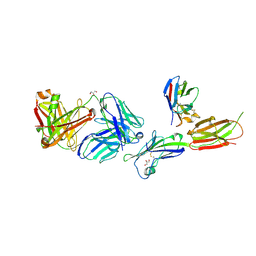 | | Crystal structure of ICOS-L in complex with Prezalumab and VNAR domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Rujas, E, Sicard, T, Julien, J.P. | | Deposit date: | 2020-05-23 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural characterization of the ICOS/ICOS-L immune complex reveals high molecular mimicry by therapeutic antibodies.
Nat Commun, 11, 2020
|
|
6X4G
 
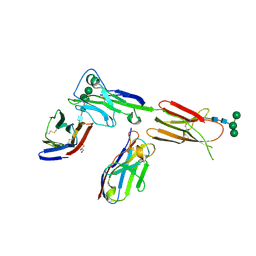 | | Crystal structure of ICOS in complex with ICOS-L and an anti ICOS-L VNAR domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ICOS ligand, ... | | Authors: | Rujas, E, Sicard, T, Julien, J.P. | | Deposit date: | 2020-05-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural characterization of the ICOS/ICOS-L immune complex reveals high molecular mimicry by therapeutic antibodies.
Nat Commun, 11, 2020
|
|
5X08
 
 | | Crystal structure of broadly neutralizing anti-HIV-1 antibody 4E10, mutant Npro, with peptide bound | | Descriptor: | ACETATE ION, CHLORIDE ION, Envelope glycoprotein gp160, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Functional Contacts between MPER and the Anti-HIV-1 Broadly Neutralizing Antibody 4E10 Extend into the Core of the Membrane
J. Mol. Biol., 429, 2017
|
|
7K9Z
 
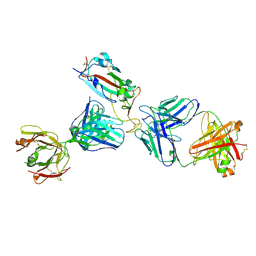 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with the Fab fragments of neutralizing antibodies 298 and 52 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 298 Fab Heavy Chain, 298 Fab Light Chain, ... | | Authors: | Newton, J.C, Kucharska, I, Rujas, E, Cui, H, Julien, J.P. | | Deposit date: | 2020-09-29 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers.
Nat Commun, 12, 2021
|
|
5GHW
 
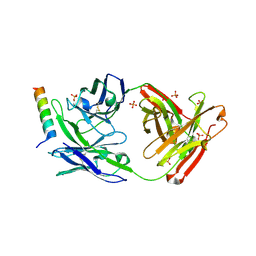 | | Crystal structure of broad neutralizing antibody 10E8 with long epitope bound | | Descriptor: | Endogenous retrovirus group K member 8 Env polyprotein, FAB 10E8 HEAVY CHAIN, FAB 10E8 LIGHT CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Morante, K, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2016-06-21 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface
Sci Rep, 6, 2016
|
|
7EKB
 
 | | Crystal structure of 4E10 modified with pyrene acetamide | | Descriptor: | ACETATE ION, Fab region of the heavy chain of broadly neutralizing antibody anti-HIV-1 4E10, Fab region of the light chain of the broadly neutralizing anti-HIV-1 antibody 4E10, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Focal accumulation of aromaticity at the CDRH3 loop mitigates 4E10 polyreactivity without altering its HIV neutralization profile.
Iscience, 24, 2021
|
|
7EKK
 
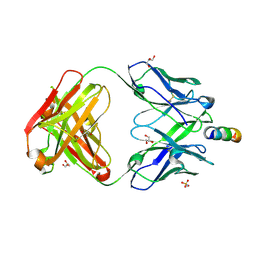 | | Anti-HIV-1 broadly neutralizing antibody delta-loop 4E10 modified with pyrene acetamide | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Focal accumulation of aromaticity at the CDRH3 loop mitigates 4E10 polyreactivity without altering its HIV neutralization profile.
Iscience, 24, 2021
|
|
5CIN
 
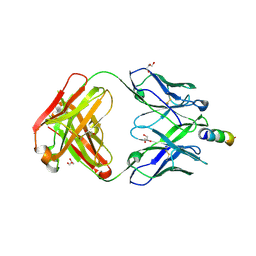 | | Crystal Structure of non-neutralizing version of 4E10 (DeltaLoop) with epitope bound | | Descriptor: | CHLORIDE ION, FAB 4E10 HEAVY CHAIN, FAB 4E10 LIGHT CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
5CIP
 
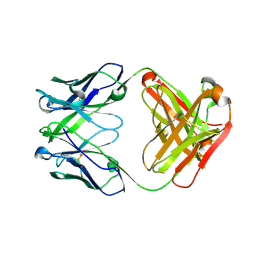 | | Crystal Structure of Unbound 4E10 | | Descriptor: | FAB 4E10 HEAVY CHAIN, FAB 4E10 LIGHT CHAIN | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
5CIL
 
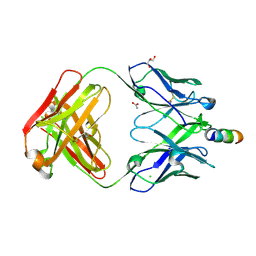 | | Crystal Structure of non-neutralizing version of 4E10 (WDWD) with epitope bound | | Descriptor: | ACETATE ION, CHLORIDE ION, FAB 4E10 HEAVY CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2015-07-13 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Thermodynamic Basis of Epitope Binding by Neutralizing and Nonneutralizing Forms of the Anti-HIV-1 Antibody 4E10
J.Virol., 89, 2015
|
|
2NCS
 
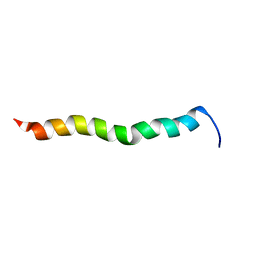 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
2NCT
 
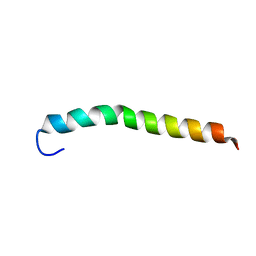 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
