6CB8
 
 | |
4DQN
 
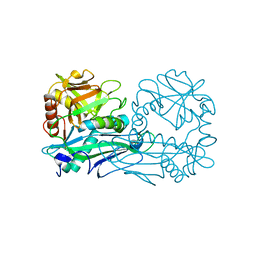 | |
4FBD
 
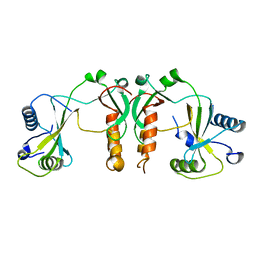 | | 2.35 Angstrom Crystal Structure of Conserved Hypothetical Protein from Toxoplasma gondii ME49. | | Descriptor: | Putative uncharacterized protein | | Authors: | Minasov, G, Ruan, J, Wawrzak, Z, Shuvalova, L, Ngo, H, Knoll, L, Milligan-Myhre, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom Crystal Structure of Conserved Hypothetical Protein from Toxoplasma gondii ME49.
TO BE PUBLISHED
|
|
5BXI
 
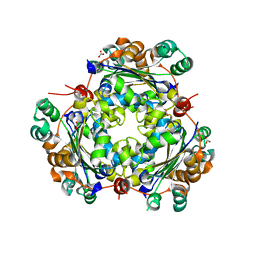 | | 1.7 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii with Tyrosine of Tag Bound to Active Site | | Descriptor: | BICARBONATE ION, DI(HYDROXYETHYL)ETHER, Nucleoside diphosphate kinase | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-06-09 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4NML
 
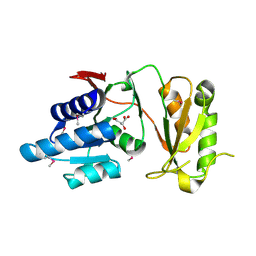 | | 2.60 Angstrom resolution crystal structure of putative ribose 5-phosphate isomerase from Toxoplasma gondii ME49 in complex with DL-Malic acid | | Descriptor: | CHLORIDE ION, D-MALATE, Ribulose 5-phosphate isomerase | | Authors: | Halavaty, A.S, Dubrovska, I, Flores, K, Shanmugam, D, Shuvalova, L, Roos, D, Ruan, J, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4NOG
 
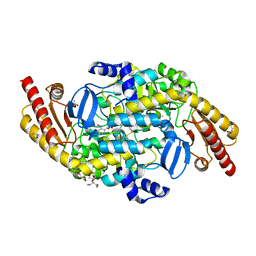 | | Crystal structure of a putative ornithine aminotransferase from Toxoplasma gondii ME49 in complex with pyrodoxal-5'-phosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Halavaty, A, Ruan, J, Shuvalova, L, Flores, K, Dubrovska, I, Ngo, H, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4NU7
 
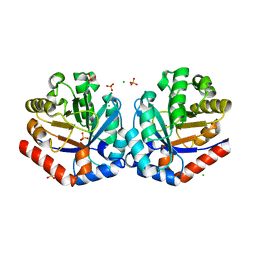 | | 2.05 Angstrom Crystal Structure of Ribulose-phosphate 3-epimerase from Toxoplasma gondii. | | Descriptor: | CHLORIDE ION, Ribulose-phosphate 3-epimerase, SULFATE ION, ... | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-03 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4O0N
 
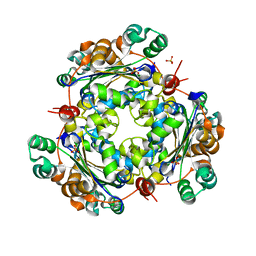 | | 2.4 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii. | | Descriptor: | Nucleoside diphosphate kinase, SULFATE ION | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-13 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4IR8
 
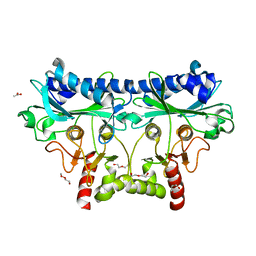 | | 1.85 Angstrom Crystal Structure of Putative Sedoheptulose-1,7 bisphosphatase from Toxoplasma gondii | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Sedoheptulose-1,7 bisphosphatase, ... | | Authors: | Minasov, G, Ruan, J, Wawrzak, Z, Halavaty, A, Shuvalova, L, Harb, O.S, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Crystal Structure of Putative Sedoheptulose-1,7 bisphosphatase from Toxoplasma gondii.
TO BE PUBLISHED
|
|
4MJZ
 
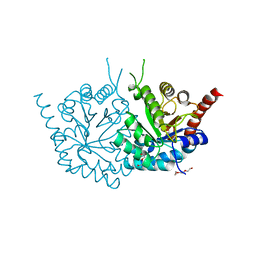 | | 2.75 Angstrom Resolution Crystal Structure of Putative Orotidine-monophosphate-decarboxylase from Toxoplasma gondii. | | Descriptor: | DI(HYDROXYETHYL)ETHER, NITRATE ION, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Minasov, G, Wawrzak, Z, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of Putative Orotidine-monophosphate-decarboxylase from Toxoplasma gondii.
TO BE PUBLISHED
|
|
8ET2
 
 | | CryoEM structure of the GSDMB pore | | Descriptor: | Isoform 1 of Gasdermin-B | | Authors: | Wang, C, Ruan, J. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.96 Å) | | Cite: | Structural basis for GSDMB pore formation and its targeting by IpaH7.8.
Nature, 616, 2023
|
|
8ET1
 
 | |
3JBL
 
 | | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Zhang, L, Chen, S, Ruan, J, Wu, J, Tong, A.B, Yin, Q, Li, Y, David, L, Lu, A, Wang, W.L, Marks, C, Ouyang, Q, Zhang, X, Mao, Y, Wu, H. | | Deposit date: | 2015-09-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization.
Science, 350, 2015
|
|
8EFP
 
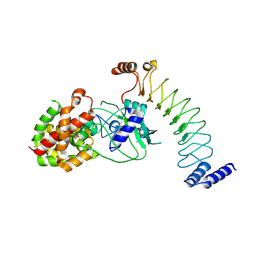 | |
3J63
 
 | | Unified assembly mechanism of ASC-dependent inflammasomes | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD | | Authors: | Lu, A, Magupalli, V.G, Ruan, J, Yin, Q, Atianand, M.K, Vos, M, Schroder, G.F, Fitzgerald, K.A, Wu, H, Egelman, E.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes.
Cell(Cambridge,Mass.), 156, 2014
|
|
4ODI
 
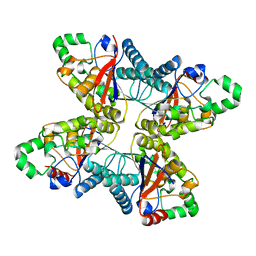 | | 2.6 Angstrom Crystal Structure of Putative Phosphoglycerate Mutase 1 from Toxoplasma gondii | | Descriptor: | Phosphoglycerate mutase PGMII, SODIUM ION | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-10 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
5H8O
 
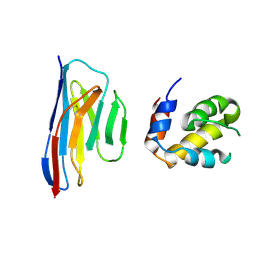 | | Crystal structure of an ASC-binding nanobody in complex with the CARD domain of ASC | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD, VHH nanobody | | Authors: | Lu, A, Schmidt, F.I, Ruan, J, Tang, C, Wu, H, Ploegh, H.L. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.206 Å) | | Cite: | A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly.
J.Exp.Med., 213, 2016
|
|
5I4C
 
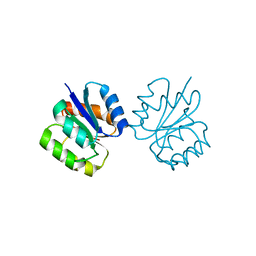 | | Crystal structure of non-phosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli | | Descriptor: | Transcriptional regulatory protein RcsB | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Ruan, J, Pshenychnyi, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of nonphosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli.
Protein Sci., 25, 2016
|
|
5H8D
 
 | | Crystal structure of an ASC binding nanobody | | Descriptor: | VHH nanobody | | Authors: | Lu, A, Schmidt, F.I, Ruan, J, Tang, C, Wu, H, Ploegh, H.L. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly.
J.Exp.Med., 213, 2016
|
|
7KEU
 
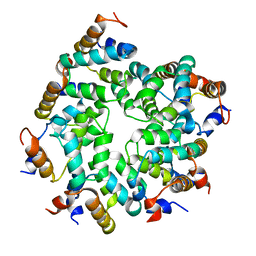 | | Cryo-EM structure of the Caspase-1-CARD:ASC-CARD octamer | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD, Caspase-1 | | Authors: | Hollingsworth, L.R, David, L, Li, Y, Ruan, J, Wu, H. | | Deposit date: | 2020-10-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
6VFE
 
 | | Gasdermin D pore | | Descriptor: | Gasdermin-D, N-terminal | | Authors: | Xia, S, Ruan, J, Wu, H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Gasdermin D pore structure reveals preferential release of mature interleukin-1.
Nature, 593, 2021
|
|
3R93
 
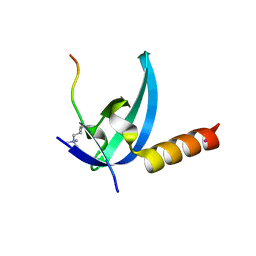 | | Crystal structure of the chromo domain of M-phase phosphoprotein 8 bound to H3K9Me3 peptide | | Descriptor: | H3K9Me3 peptide, M-phase phosphoprotein 8, UNKNOWN ATOM OR ION | | Authors: | Li, J, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
3QYQ
 
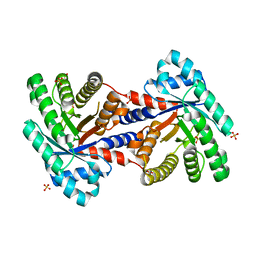 | | 1.8 Angstrom resolution crystal structure of a putative deoxyribose-phosphate aldolase from Toxoplasma gondii ME49 | | Descriptor: | Deoxyribose-phosphate aldolase, putative, SULFATE ION, ... | | Authors: | Halavaty, A.S, Ruan, J, Minasov, G, Shuvalova, L, Ueno, A, Igarashi, M, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional divergence of the aldolase fold in Toxoplasma gondii.
J.Mol.Biol., 427, 2015
|
|
3MP6
 
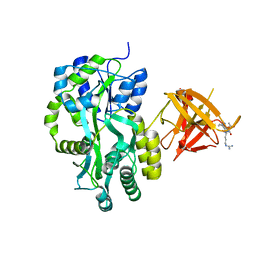 | | Complex Structure of Sgf29 and dimethylated H3K4 | | Descriptor: | H3K4me2 peptide, Maltose-binding periplasmic protein,LINKER,SAGA-associated factor 29, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, J, Wu, M, Ruan, J, Zang, J. | | Deposit date: | 2010-04-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
3MP8
 
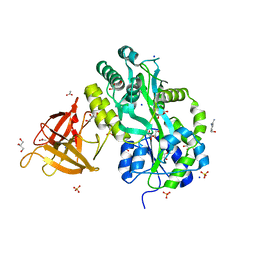 | | Crystal structure of Sgf29 tudor domain | | Descriptor: | 4-(HYDROXYMETHYL)BENZAMIDINE, ACETIC ACID, GLYCEROL, ... | | Authors: | Li, J, Wu, M, Ruan, J, Zang, J. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
