4AN1
 
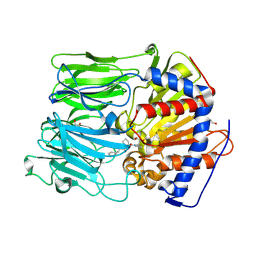 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-4 | | Descriptor: | (2S)-N-benzyl-2-({(2S)-2-[(1R)-1,2-dihydroxyethyl]pyrrolidin-1-yl}carbonyl)pyrrolidine-1-carboxamide, GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Dynamics, Crystallography and Mutagenesis Studies on the Substrate Gating Mechanism of Prolyl Oligopeptidase.
Biochimie, 94, 2012
|
|
4AN0
 
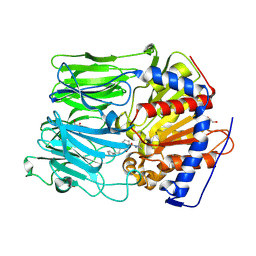 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-3 | | Descriptor: | (2S)-1-[1-(4-phenylbutanoyl)-L-prolyl]pyrrolidine-2-carbonitrile, GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular dynamics, crystallography and mutagenesis studies on the substrate gating mechanism of prolyl oligopeptidase.
Biochimie, 94, 2012
|
|
4AMY
 
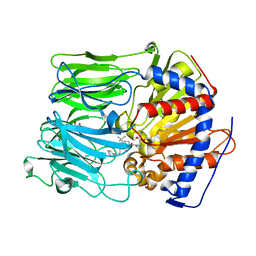 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-1 | | Descriptor: | 1-[(2R,5S)-2-tert-butyl-5-({(2S)-2-[(1R)-1,2-dihydroxyethyl]pyrrolidin-1-yl}carbonyl)pyrrolidin-1-yl]-4-phenylbutan-1-one, GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Dynamics, Crystallography and Mutagenesis Studies on the Substrate Gating Mechanism of Prolyl Oligopeptidase.
Biochimie, 94, 2012
|
|
4AMZ
 
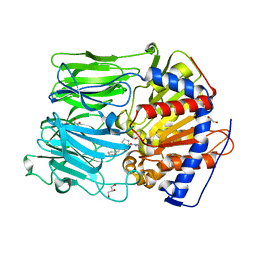 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-2 | | Descriptor: | (5R)-N-benzyl-5-({(2S)-2-[(1R)-1,2-dihydroxyethyl]pyrrolidin-1-yl}carbonyl)cyclopent-1-ene-1-carboxamide, GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Dynamics, Crystallography and Mutagenesis Studies on the Substrate Gating Mechanism of Prolyl Oligopeptidase.
Biochimie, 94, 2012
|
|
4UFC
 
 | | Crystal structure of the GH95 enzyme BACOVA_03438 | | Descriptor: | CACODYLATE ION, CALCIUM ION, GH95, ... | | Authors: | Rogowski, A, Briggs, J.A, Mortimer, J.C, Tryfona, T, Terrapon, N, Lowe, E.C, Basle, A, Morland, C, Day, A.M, Zheng, H, Rogers, T.E, Thompson, P, Hawkins, A.R, Yadav, M.P, Henrissat, B, Martens, E.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Glycan Complexity Dictates Microbial Resource Allocation in the Large Intestine.
Nat.Commun., 6, 2015
|
|
6JNA
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JN9
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JNC
 
 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
6JND
 
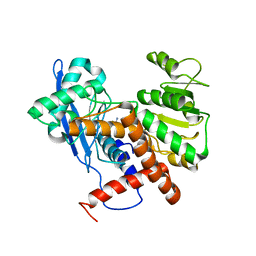 | | Cryo-EM structure of glutamate dehydrogenase from Thermococcus profundus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate dehydrogenase | | Authors: | Oide, M, Kato, T, Oroguchi, T, Nakasako, M. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Energy landscape of domain motion in glutamate dehydrogenase deduced from cryo-electron microscopy.
Febs J., 287, 2020
|
|
8TOO
 
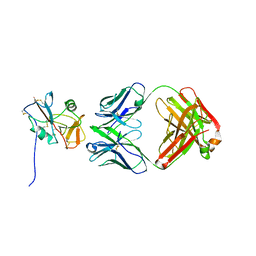 | | Crystal structure of Epstein-Barr virus gp42 in complex with antibody 4C12 | | Descriptor: | 4C12 heavy chain, 4C12 light chain, Glycoprotein 42 | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
8TNN
 
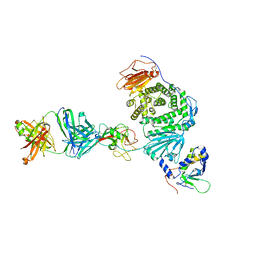 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with gp42 antibody A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A10 heavy chain, A10 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
3ZMR
 
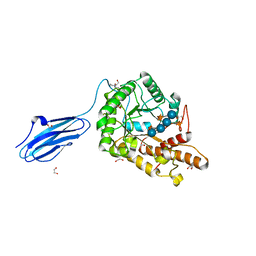 | | Bacteroides ovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CELLULASE (GLYCOSYL HYDROLASE FAMILY 5), ... | | Authors: | Larsbrink, J, Rogers, T.E, Hemsworth, G.R, McKee, L.S, Spadiut, O, Klinter, S, Pudlo, N.A, Urs, K, Kelly, A.G, Cederholm, S.N, Davies, G.J, Martens, E.C, Brumer, H. | | Deposit date: | 2013-02-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Discrete Genetic Locus Confers Xyloglucan Metabolism in Select Human Gut Bacteroidetes
Nature, 506, 2014
|
|
8TNT
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with antibodies F-2-1 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 heavy chain, 769C2 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
5NBP
 
 | | Bacteroides ovatus mixed linkage glucan PUL (MLGUL) GH16 in complex with G4G4G3G Product | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 16, ... | | Authors: | Hemsworth, G.R, Tamura, K, Dejean, G, Rogers, T.E, Pudlo, N.A, Urs, K, Jain, N, Martens, E.C, Brumer, H, Davies, G.J. | | Deposit date: | 2017-03-02 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism by which Prominent Human Gut Bacteroidetes Utilize Mixed-Linkage Beta-Glucans, Major Health-Promoting Cereal Polysaccharides.
Cell Rep, 21, 2017
|
|
5NBO
 
 | | Bacteroides ovatus mixed linkage glucan PUL (MLGUL) GH16 | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase family 16 | | Authors: | Hemsworth, G.R, Tamura, K, Dejean, G, Rogers, T.E, Pudlo, N.A, Urs, K, Jain, N, Martens, E.C, Brumer, H, Davies, G.J. | | Deposit date: | 2017-03-02 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism by which Prominent Human Gut Bacteroidetes Utilize Mixed-Linkage Beta-Glucans, Major Health-Promoting Cereal Polysaccharides.
Cell Rep, 21, 2017
|
|
3ZBO
 
 | | A new family of proteins related to the HEAT-like repeat DNA glycosylases with affinity for branched DNA structures | | Descriptor: | ALKF, CHLORIDE ION | | Authors: | Backe, P.H, Simm, R, Laerdahl, J.K, Dalhus, B, Fagerlund, A, Okstad, O.A, Rognes, T, Alseth, I, Kolsto, A.-B, Bjoras, M. | | Deposit date: | 2012-11-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A New Family of Proteins Related to the Heat-Like Repeat DNA Glycosylases with Affinity for Branched DNA Structures.
J.Struct.Biol., 183, 2013
|
|
7KCR
 
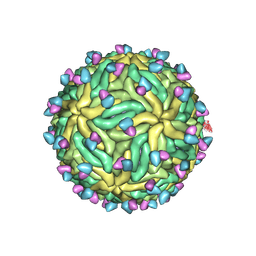 | | Cryo-EM structure of Zika virus in complex with E protein cross-linking human monoclonal antibody ADI30056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADI30056 Fab heavy chain variable region, ADI30056 Fab light chain variable region, ... | | Authors: | Sevvana, M, Rogers, T.F, Miller, A.S, Long, F, Klose, T, Beutler, N, Lai, Y.C, Parren, M, Walker, L.M, Buda, G, Burton, D.R, Rossmann, M.G, Kuhn, R.J. | | Deposit date: | 2020-10-07 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Zika Virus Specific Neutralization in Subsequent Flavivirus Infections.
Viruses, 12, 2020
|
|
